Efficient low-temperature wastewater treatment by Pseudomonas zhanjiangensis sp. nov.: a novel cold-tolerant bacterium isolated from mangrove sediment
- PMID: 39545239
- PMCID: PMC11560893
- DOI: 10.3389/fmicb.2024.1491174
Efficient low-temperature wastewater treatment by Pseudomonas zhanjiangensis sp. nov.: a novel cold-tolerant bacterium isolated from mangrove sediment
Abstract
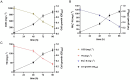
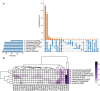
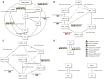
A novel heterotrophic, cold-tolerant bacterium, designated Pseudomonas zhanjiangensis 25A3ET, was isolated from mangrove sediment and demonstrated excellent efficiency in cold wastewater treatment. Phylogenetic analysis based on 16S rRNA gene sequences positioned strain 25A3ET within the genus Pseudomonas, showing the highest similarity (98.7%) with Pseudomonas kurunegalensis LMG 32023T. Digital DNA-DNA hybridization (dDDH) and average nucleotide identity (ANI) values were below the species delineation thresholds (70% for dDDH, 95% for ANI), indicating that strain 25A3ET represents a novel species. This strain demonstrated high efficiency in removing nitrogen (N) and organic pollutants under low-temperature conditions. Specifically, it achieved 72.9% removal of chemical oxygen demand (COD), 70.6% removal of ammoniacal nitrogen (NH4 +-N), and 69.1% removal of total nitrogen (TN) after 96 h at 10°C. Genomic analysis identified key genes associated with cold adaptation, nitrogen removal and organic matter degradation. These findings indicate that Pseudomonas zhanjiangensis 25A3ET holds significant potential for application in cold temperature wastewater treatment, offering a promising solution for environmental remediation in regions with low ambient temperatures.
Keywords: Pseudomonas zhanjiangensis; cold tolerant; comparative genomic analysis; nitrogen removal; wastewater treatment.
Copyright © 2024 Li, Hu, Ni, Ni, Li, Xue and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Isolation and characterization of a cold-tolerant heterotrophic nitrification-aerobic denitrification bacterium and evaluation of its nitrogen-removal efficiency.Environ Res. 2024 Feb 1;242:117674. doi: 10.1016/j.envres.2023.117674. Epub 2023 Nov 28. Environ Res. 2024. PMID: 38029814
-
Pseudomonas nunensis sp. nov. isolated from a suppressive potato field in Greenland.Int J Syst Evol Microbiol. 2023 Feb;73(2). doi: 10.1099/ijsem.0.005700. Int J Syst Evol Microbiol. 2023. PMID: 36749687
-
Pseudomonas citri sp. nov., a potential novel plant growth promoting bacterium isolated from rhizosphere soil of citrus.Antonie Van Leeuwenhoek. 2023 Mar;116(3):281-289. doi: 10.1007/s10482-022-01803-y. Epub 2023 Jan 3. Antonie Van Leeuwenhoek. 2023. PMID: 36596938
-
Caenimonas sedimenti sp. nov., Isolated from Sediment of the Wastewater Outlet of an Agricultural Chemical Plant.Curr Microbiol. 2020 Nov;77(11):3767-3772. doi: 10.1007/s00284-020-02145-6. Epub 2020 Aug 3. Curr Microbiol. 2020. PMID: 32748162 Review.
-
Pseudomonas pratensis sp. nov., Isolated from Grassland Soil from Inner Mongolia, China.Curr Microbiol. 2021 Feb;78(2):789-795. doi: 10.1007/s00284-020-02296-6. Epub 2021 Jan 2. Curr Microbiol. 2021. PMID: 33389060 Review.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

