A multidimensional assessment of in-host fitness costs of drug resistance in the opportunistic fungal pathogen Candida glabrata
- PMID: 39545363
- PMCID: PMC11631428
- DOI: 10.1093/femsyr/foae035
A multidimensional assessment of in-host fitness costs of drug resistance in the opportunistic fungal pathogen Candida glabrata
Abstract
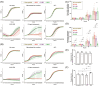
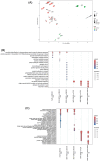
Drug-resistant microbes typically carry mutations in genes involved in critical cellular functions and may therefore be less fit under drug-free conditions than susceptible strains. Candida glabrata is a prevalent opportunistic yeast pathogen with a high rate of fluconazole resistance (FLZR), echinocandin resistance (ECR), and multidrug resistance (MDR) relative to other Candida. However, the fitness of C. glabrata MDR isolates, particularly in the host, is poorly characterized, and studies of FLZR isolate fitness have produced contradictory findings. Two important host niches for C. glabrata are macrophages, in which it survives and proliferates, and the gut. Herein, we used a collection of clinical and lab-derived C. glabrata isolates to show that FLZR C. glabrata isolates are less fit inside macrophages than susceptible isolates and that this fitness cost is reversed by acquiring ECR mutations. Interestingly, dual-RNAseq revealed that macrophages infected with drug-resistant isolates mount an inflammatory response whereas intracellular drug-resistant cells downregulate processes required for in-host adaptation. Furthermore, drug-resistant isolates were outcompeted by their susceptible counterparts during gut colonization and in infected kidneys, while showing comparable fitness in the spleen. Collectively, our study shows that macrophage-rich organs, such as the spleen, favor the retention of MDR isolates of C. glabrata.
Keywords: Candida glabrata; echinocandin resistant; fitness cost; fluconazole resistant; gut colonization; intracellular replication; multidrug resistant; systemic infection.
© The Author(s) 2024. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
None declared.
Figures





References
-
- Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance?. Nat Rev Microbiol. 2010;8:260–71. - PubMed
-
- Arason VA, Gunnlaugsson A, Sigurdsson JA et al. Clonal spread of resistant pneumococci despite diminished antimicrobial use. Microb Drug Resist. 2002;8:187–92. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

