Defects in meiosis I contribute to the genesis of androgenetic hydatidiform moles
- PMID: 39545410
- PMCID: PMC11563684
- DOI: 10.1172/JCI170669
Defects in meiosis I contribute to the genesis of androgenetic hydatidiform moles
Abstract
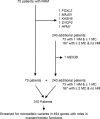
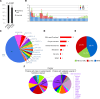
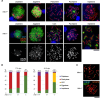
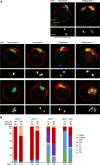
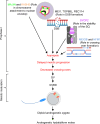
To identify novel genes responsible for recurrent hydatidiform moles (HMs), we performed exome sequencing on 75 unrelated patients who were negative for mutations in the known genes. We identified biallelic deleterious variants in 6 genes, FOXL2, MAJIN, KASH5, SYCP2, MEIOB, and HFM1, in patients with androgenetic HMs, including a familial case of 3 affected members. Five of these genes are essential for meiosis I, and their deficiencies lead to premature ovarian insufficiency. Advanced maternal age is the strongest risk factor for sporadic androgenetic HM, which affects 1 in every 600 pregnancies. We studied Hfm1-/- female mice and found that these mice lost all their oocytes before puberty but retained some at younger ages. Oocytes from Hfm1-/- mice initiated meiotic maturation and extruded the first polar bodies in culture; however, their meiotic spindles were often positioned parallel, instead of perpendicular, to the ooplasmic membrane at telophase I, and some oocytes extruded the entire spindle with all the chromosomes into the polar bodies at metaphase II, a mechanism we previously reported in Mei1-/- oocytes. The occurrence of a common mechanism in two mouse models argues in favor of its plausibility at the origin of androgenetic HM formation in humans.
Keywords: Fertility; Genetics; Monogenic diseases; Reproductive biology.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

