Gut microbiota diversity is prognostic and associated with benefit from chemo-immunotherapy in metastatic triple-negative breast cancer
- PMID: 39545921
- PMCID: PMC11977656
- DOI: 10.1002/1878-0261.13760
Gut microbiota diversity is prognostic and associated with benefit from chemo-immunotherapy in metastatic triple-negative breast cancer
Abstract
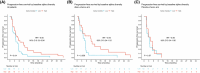
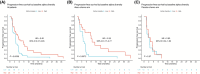
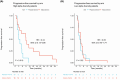
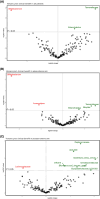
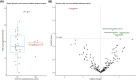
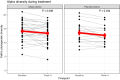
The gut microbiota influences multiple aspects of human health and disease. Several studies have indicated an association between the gut microbiota and response to immune checkpoint inhibitors in various cancers, but there is scarce data from breast cancer. The randomized ALICE trial demonstrated improved progression-free survival (PFS) from adding the programmed cell death 1 ligand 1 (PD-L1) inhibitor atezolizumab (atezo) to immunomodulating chemotherapy (chemo) in metastatic triple-negative breast cancer (mTNBC), even for PD-L1negative disease. Herein, we investigated the microbiota composition and dynamics in the ALICE patients and their association with clinical outcome, by analyzing fecal samples collected at baseline and after 8 weeks. We applied 16S (V3-V4) rRNA sequencing to characterize the diversity and taxonomic composition. Kaplan-Meier and Cox proportional hazard models were used for time-to-event analyses. We found that high alpha diversity by Faith's phylogenetic diversity (PD) at baseline was associated with prolonged PFS in the total study population and in the atezo-chemo arm, but not in the placebo-chemo arm. Moreover, Faith's PD appeared to be predictive of benefit from atezolizumab. Patients with high Faith's PD exhibited a PFS hazard ratio of 0.34 (P = 0.018) in favor of the atezo-chemo arm, compared to 0.83 (P = 0.62) in the low Faith's PD group. Faith's PD was significantly reduced during treatment. At baseline, Bifidobacterium was significantly overrepresented in patients without clinical benefit in the atezo-chemo arm, but not in the placebo-chemo arm. These findings suggest that alpha diversity by Faith's PD should be further investigated as a prognostic and predictive biomarker in patients with mTNBC receiving chemo-immunotherapy.
Keywords: alpha diversity; biomarker; gut microbiota; immunotherapy; triple‐negative breast cancer.
© 2024 The Author(s). Molecular Oncology published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
J.A.K. has the last five years received research support from Bristol Myers Squibb, F. Hoffmann‐La Roche, NanoString and NEC OncoImmunity and has previously received advisory board/lecture honoraria from pharmaceutical companies, including Roche. JRH has received grants from Biogen and lecture honoraria from Amgen, Roche and Novartis. The other authors declare no conflicts of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

