Microglia contribute to the production of the amyloidogenic ABri peptide in familial British dementia
- PMID: 39546024
- PMCID: PMC11568029
- DOI: 10.1007/s00401-024-02820-z
Microglia contribute to the production of the amyloidogenic ABri peptide in familial British dementia
Abstract
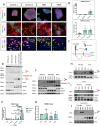
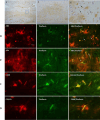
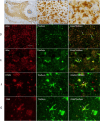
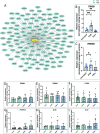
Mutations in ITM2B cause familial British, Danish, Chinese, and Korean dementias. In familial British dementia (FBD), a mutation in the stop codon of the ITM2B gene (also known as BRI2) causes a C-terminal cleavage fragment of the ITM2B/BRI2 protein to be extended by 11 amino acids. This fragment, termed amyloid-Bri (ABri), is highly insoluble and forms extracellular plaques in the brain. ABri plaques are accompanied by tau pathology, neuronal cell death and progressive dementia, with striking parallels to the aetiology and pathogenesis of Alzheimer's disease. The molecular mechanisms underpinning FBD are ill-defined. Using patient-derived induced pluripotent stem cells, we show that expression of ITM2B/BRI2 is 34-fold higher in microglia than neurons and 15-fold higher in microglia compared with astrocytes. This cell-specific enrichment is supported by expression data from both mouse and human brain tissue. ITM2B/BRI2 protein levels are higher in iPSC-microglia compared with neurons and astrocytes. The ABri peptide was detected in patient iPSC-derived microglial lysates and conditioned media but was undetectable in patient-derived neurons and control microglia. The pathological examination of post-mortem tissue supports the presence of ABri in microglia that are in proximity to pre-amyloid deposits. Finally, gene co-expression analysis supports a role for ITM2B/BRI2 in disease-associated microglial responses. These data demonstrate that microglia are major contributors to the production of amyloid forming peptides in FBD, potentially acting as instigators of neurodegeneration. Additionally, these data also suggest ITM2B/BRI2 may be part of a microglial response to disease, motivating further investigations of its role in microglial activation. These data have implications for our understanding of the role of microglia and the innate immune response in the pathogenesis of FBD and other neurodegenerative dementias including Alzheimer's disease.
Keywords: Alzheimer’s disease; Amyloid; Dementia; Familial British dementia; Microglia; iPSC.
© 2024. The Author(s).
Conflict of interest statement
Figures
Update of
-
Microglia produce the amyloidogenic ABri peptide in familial British dementia.bioRxiv [Preprint]. 2023 Jun 28:2023.06.27.546552. doi: 10.1101/2023.06.27.546552. bioRxiv. 2023. Update in: Acta Neuropathol. 2024 Nov 15;148(1):65. doi: 10.1007/s00401-024-02820-z. PMID: 37425748 Free PMC article. Updated. Preprint.
References
-
- Audo I, Bujakowska K, Orhan E, El Shamieh S, Sennlaub F, Guillonneau X et al (2014) The familial dementia gene revisited: a missense mutation revealed by whole-exome sequencing identifies ITM2B as a candidate gene underlying a novel autosomal dominant retinal dystrophy in a large family. Hum Mol Genet 23:491–501. 10.1093/hmg/ddt439 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

