A High-Fidelity Computational Model for Predicting Blood Cell Trafficking and 3D Capillary Hemodynamics in Retinal Microvascular Networks
- PMID: 39546289
- PMCID: PMC11580294
- DOI: 10.1167/iovs.65.13.37
A High-Fidelity Computational Model for Predicting Blood Cell Trafficking and 3D Capillary Hemodynamics in Retinal Microvascular Networks
Abstract
Purpose: To present a first principle-based, high-fidelity computational model for predicting full three-dimensional (3D) and time-resolved retinal microvascular hemodynamics taking into consideration the flow and deformation of individual blood cells.
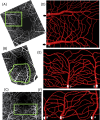
Methods: The computational model is a 3D fluid-structure interaction model based on combined finite volume/finite element/immersed-boundary methods. Three in silico microvascular networks are built from high-resolution in vivo motion contrast images of the superficial capillary plexus in the parafoveal region of the human retina. The maximum tissue area represented in the model is approximately 500 × 500 µm2, and vessel lumen diameters ranged from 5.5 to 25 µm covering capillaries, arterioles, and venules. Blood is modeled as a suspension of individual blood cells, namely, erythrocytes (RBC), leukocytes (WBC), and platelets in plasma. An accurate and detailed biophysical modeling of each blood cell and their flow-induced deformation is considered. A physiological, pulsatile boundary condition corresponding to an average cardiac cycle of 0.9 second is used.
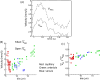
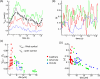
Results: Detailed quantitative data and analysis of 3D retinal microvascular hemodynamics are presented, and their relationship to RBC flow dynamics is illustrated. Blood velocity is shown to have temporal oscillations superimposed on the background pulsatile variation, which arise because of the way RBCs partition at vascular junctions, causing repeated clogging and unclogging of vessels. Temporal variations in RBC velocity and hematocrit are anti-correlated in a given vessel, but their time-averaged distributions are positively correlated across the network. Whole blood velocity is 65% to 85% of RBC velocity, with the discrepancy related to the formation of an RBC-free region, adjacent to the vascular endothelium and typically 0.8 to 1.8 µm thick. The 3D velocity and RBC concentration profiles are shown to be oppositely skewed with respect to each other, because of the way that RBCs "hug" the apex of each bifurcation. RBC deformation is predicted to have biphasic behavior with respect to vessel diameter, with minimal cell length for vessels approximately 7 µm in diameter. The wall shear stress (WSS) exhibits a strongly 3D distribution with local regions of high value and gradient spanning a range of 10 to 80 dyn/cm2. WSS is highest where there is faster flow, greater curvature of the vessel wall, capillary bifurcations, and at locations of RBC crowding and associated thinning of the cell-free layer.
Conclusions: This study highlights the usefulness of high-fidelity cell-resolved modeling to obtain accurate and detailed 3D, time-resolved retinal hemodynamic parameters that are not readily available through noninvasive imaging approaches. The results presented are expected to complement and enhance the interpretation of in vivo data, as well as open new avenues to study retinal hemodynamics in health and disease.
Conflict of interest statement
Disclosure:
Figures








References
-
- Grunwald JE, Riva CE, Sinclair SH, Brucker AJ, Petrig BL. Retinal blood flow in health and disease. Prog Retinal Eye Res. 1989; 9: 365–393.
-
- Linsenmeier RA, Zhang HF. Retinal blood flow: physiology, methods of measurement, and implications for neurodegenerative diseases. Invest Ophthalmol Vis Sci. 2017; 58: 2486–2497.
-
- Yu D-Yi, Cringle SJ, Yu PK, et al. .. Retinal capillary perfusion: spatial and temporal heterogeneity. Prog Retinal Eye Res. 2019; 30: 23–5. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

