Repurposing of drug candidates against Epstein-Barr virus: Virtual screening, docking computations, molecular dynamics, and quantum mechanical study
- PMID: 39546470
- PMCID: PMC11567563
- DOI: 10.1371/journal.pone.0312100
Repurposing of drug candidates against Epstein-Barr virus: Virtual screening, docking computations, molecular dynamics, and quantum mechanical study
Abstract
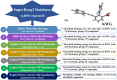
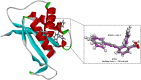
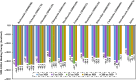
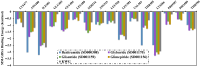
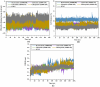
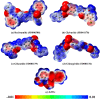
Epstein-Barr virus (EBV) was the first tumor virus identified in humans, and it is mostly linked to lymphomas and cancers of epithelial cells. Nevertheless, there is no FDA-licensed drug feasible for this ubiquitous EBV viral contagion. EBNA1 (Epstein-Barr nuclear antigen 1) plays several roles in the replication and transcriptional of latent gene expression of the EBV, making it an attractive druggable target for the treatment of EBV-related malignancies. The present study targets EBV viral reactivation and upkeep by inhibiting EBNA1 utilizing a drug-repurposing strategy. To hunt novel EBNA1 inhibitors, a SuperDRUG2 database (> 4,600 pharmaceutical ingredients) was virtually screened utilizing docking computations. In accordance with the estimated docking scores, the most promising drug candidates then underwent MDS (molecular dynamics simulations). Besides, the MM-GBSA approach was applied to estimate the binding affinities between the identified drug candidates and EBNA1. On the basis of MM-GBSA//200 ns MDS, bezitramide (SD000308), glyburide (SD001170), glisentide (SD001159), and glimepiride (SD001156) unveiled greater binding affinities towards EBNA1 compared to KWG, a reference inhibitor, with ΔGbinding values of -44.3, -44.0, -41.7, -40.2, and -32.4 kcal/mol, respectively. Per-residue decomposition analysis demonstrated that LYS477, ASN519, and LYS586 significantly interacted with the identified drug candidates within the EBNA1 binding pocket. Post-dynamic analyses also demonstrated high constancy of the identified drug candidates in complex with EBNA1 throughout 200 ns MDS. Ultimately, electrostatic potential and frontier molecular orbitals analyses were performed to estimate the chemical reactivity of the identified EBNA1 inhibitors. Considering the current outcomes, this study would be an adequate linchpin for forthcoming research associated with the inhibition of EBNA1; however, experimental assays are required to inspect the efficiency of these candidates.
Copyright: © 2024 Ibrahim et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

