MosAIC: An annotated collection of mosquito-associated bacteria with high-quality genome assemblies
- PMID: 39546548
- PMCID: PMC11633956
- DOI: 10.1371/journal.pbio.3002897
MosAIC: An annotated collection of mosquito-associated bacteria with high-quality genome assemblies
Abstract
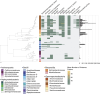
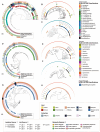
Mosquitoes transmit medically important human pathogens, including viruses like dengue virus and parasites such as Plasmodium spp., the causative agent of malaria. Mosquito microbiomes are critically important for the ability of mosquitoes to transmit disease-causing agents. However, while large collections of bacterial isolates and genomic data exist for vertebrate microbiomes, the vast majority of work in mosquitoes to date is based on 16S rRNA gene amplicon data that provides limited taxonomic resolution and no functional information. To address this gap and facilitate future studies using experimental microbiome manipulations, we generated a bacterial Mosquito-Associated Isolate Collection (MosAIC) consisting of 392 bacterial isolates with extensive metadata and high-quality draft genome assemblies that are publicly available, both isolates and sequence data, for use by the scientific community. MosAIC encompasses 142 species spanning 29 bacterial families, with members of the Enterobacteriaceae comprising 40% of the collection. Phylogenomic analysis of 3 genera, Enterobacter, Serratia, and Elizabethkingia, reveal lineages of mosquito-associated bacteria isolated from different mosquito species in multiple laboratories. Investigation into species' pangenomes further reveals clusters of genes specific to these lineages, which are of interest for future work to test for functions connected to mosquito host association. Altogether, we describe the generation of a physical collection of mosquito-associated bacterial isolates, their genomic data, and analyses of selected groups in context of genome data from closely related isolates, providing a unique, highly valuable resource for research on bacterial colonisation and adaptation within mosquito hosts. Future efforts will expand the collection to include broader geographic and host species representation, especially from individuals collected from field populations, as well as other mosquito-associated microbes, including fungi, archaea, and protozoa.
Copyright: © 2024 Foo et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

