JmjC domain-containing histone demethylase gene family in Chinese cabbage: Genome-wide identification and expressional profiling
- PMID: 39546552
- PMCID: PMC11567544
- DOI: 10.1371/journal.pone.0312798
JmjC domain-containing histone demethylase gene family in Chinese cabbage: Genome-wide identification and expressional profiling
Abstract
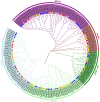
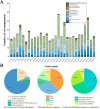
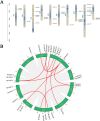
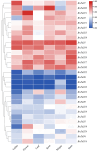
The Jumonji C (JmjC) structural domain-containing gene family plays essential roles in stress responses. However, descriptions of this family in Brassica rapa ssp. pekinensis (Chinese cabbage) are still scarce. In this study, we identified 29 members of the BrJMJ gene family, with cis-acting elements related to light, low temperature, anaerobic conditions, and phytohormone responses. Most BrJMJs were highly expressed in the siliques and flowers, suggesting that histone demethylation may play a crucial role in reproductive organ development. The expression of BrJMJ1, BrJMJ2, BrJMJ5, BrJMJ13, BrJMJ21 and BrJMJ24 gradually increased with higher Cd concentration under Cd stress, while BrJMJ4 and BrJMJ29 could be induced by osmotic, salt, cold, and heat stress. These results demonstrate that BrJMJs are responsive to abiotic stress and support future analysis of their biological functions.
Copyright: © 2024 Yin et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Characterization and expression profiling of MYB transcription factors against stresses and during male organ development in Chinese cabbage (Brassica rapa ssp. pekinensis).Plant Physiol Biochem. 2016 Jul;104:200-15. doi: 10.1016/j.plaphy.2016.03.021. Epub 2016 Mar 18. Plant Physiol Biochem. 2016. PMID: 27038155
-
Genome-Wide Investigation and Expression Analysis of K+-Transport-Related Gene Families in Chinese Cabbage (Brassica rapa ssp. pekinensis).Biochem Genet. 2021 Feb;59(1):256-282. doi: 10.1007/s10528-020-10004-z. Epub 2020 Sep 29. Biochem Genet. 2021. PMID: 32990910
-
Genome-wide characterization and expression profiling of PDI family gene reveals function as abiotic and biotic stress tolerance in Chinese cabbage (Brassica rapa ssp. pekinensis).BMC Genomics. 2017 Nov 16;18(1):885. doi: 10.1186/s12864-017-4277-2. BMC Genomics. 2017. PMID: 29145809 Free PMC article.
-
Gene co-expression network analysis reveals key pathways and hub genes in Chinese cabbage (Brassica rapa L.) during vernalization.BMC Genomics. 2021 Apr 6;22(1):236. doi: 10.1186/s12864-021-07510-8. BMC Genomics. 2021. PMID: 33823810 Free PMC article. Review.
-
Epigenetic gene regulation by plant Jumonji group of histone demethylase.Biochim Biophys Acta. 2011 Aug;1809(8):421-6. doi: 10.1016/j.bbagrm.2011.03.004. Epub 2011 Mar 16. Biochim Biophys Acta. 2011. PMID: 21419882 Review.
References
-
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Timothy Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997; 389:251–260. 10.1038/38444. - PubMed
-
- Niu Y, Bai J, Zheng S. The Regulation and Function of Histone Methylation. Journal of Plant Biology. 2018; 61(6):347–357. 10.1007/s12374-018-01766.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

