Loss of XIST lncRNA unlocks stemness and cellular plasticity in ovarian cancer
- PMID: 39546568
- PMCID: PMC11588085
- DOI: 10.1073/pnas.2418096121
Loss of XIST lncRNA unlocks stemness and cellular plasticity in ovarian cancer
Abstract
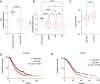
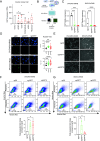
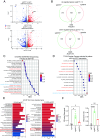
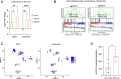
Plasticity, a key hallmark of cancer, enables cells to transition into different states, driving tumor heterogeneity. This cellular plasticity is associated with cancer progression, treatment resistance, and relapse. Cancer stem cells (CSCs) play a central role in this process, yet the molecular factors underlying cancer cell stemness remain poorly understood. In this study, we explored the role of XIST (X-inactive specific transcript) long noncoding RNA in ovarian cancer stemness and plasticity through in silico and in vitro analyses. We found that XIST is significantly down-regulated in ovarian tumors, with low XIST expression linked to a higher stemness index and lower overall survival. Knocking down XIST in ovarian cancer cells enhanced stemness, particularly increasing mesenchymal-like CSCs, and under hypoxic conditions, it promoted epithelial-like CSC markers. Our findings suggest that XIST loss leads to CSC enrichment and cellular plasticity in ovarian cancer, pointing to potential therapeutic targets for patients with low XIST expression.
Keywords: XIST; cancer stem cell; cellular plasticity; epigenetics; ovarian cancer.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

