S-adenosyl-L-methionine is the unexpected methyl donor for the methylation of mercury by the membrane-associated HgcAB complex
- PMID: 39546574
- PMCID: PMC11588087
- DOI: 10.1073/pnas.2408086121
S-adenosyl-L-methionine is the unexpected methyl donor for the methylation of mercury by the membrane-associated HgcAB complex
Abstract
Mercury (Hg) is a heavy metal that exhibits high biological toxicity. Monomethylmercury and dimethylmercury are neurotoxins and a significant environmental concern as they bioaccumulate and biomagnify within the aquatic food web. Microbial Hg methylation involves two proteins, HgcA and HgcB. Here, we show that HgcA and HgcB can be heterologously coexpressed, and the HgcAB complex can be purified. We demonstrated that HgcA is a membrane-associated cobalamin-dependent methyltransferase and HgcB is a ferredoxin-like protein containing two [4Fe-4S] clusters. Further, spectroscopic and kinetic results demonstrate that S-adenosyl-L-methionine (SAM) donates the methyl group to Hg in a two-step reaction involving a methylcob(III)alamin intermediate including Co-thiolate ligation from a conserved Cys residue. Our findings uncover a biological role for SAM in microbial Hg methylation.
Keywords: S-adenosyl-L-methionine; cobalamin; iron-sulfur; methyl mercury; methyltransferase.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures
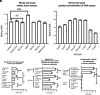
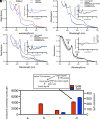



References
-
- Clarkson T. W., Human toxicology of Mercury. J. Trace Elem. Exp. Med. 11, 303–317 (1998).
-
- Parks J. M., et al. , The genetic basis for bacterial mercury methylation. Science 339, 1332–1335 (2013). - PubMed
-
- Gilmour C. C., et al. , Mercury methylation by novel microorganisms from new environments. Environ. Sci. Technol. 47, 11810–11820 (2013). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

