The HDAC6 inhibitor AVS100 (SS208) induces a pro-inflammatory tumor microenvironment and potentiates immunotherapy
- PMID: 39546602
- PMCID: PMC11566997
- DOI: 10.1126/sciadv.adp3687
The HDAC6 inhibitor AVS100 (SS208) induces a pro-inflammatory tumor microenvironment and potentiates immunotherapy
Abstract
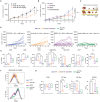
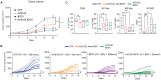
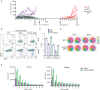
Histone deacetylase 6 (HDAC6) inhibition is associated with an increased pro-inflammatory tumor microenvironment and antitumoral immune responses. Here, we show that the HDAC6 inhibitor AVS100 (SS208) had an antitumoral effect in SM1 melanoma and CT26 colon cancer models and increased the efficacy of anti-programmed cell death protein 1 treatment, leading to complete remission in melanoma and increased response in colon cancer. AVS100 treatment increased pro-inflammatory tumor-infiltrating macrophages and CD8 effector T cells with an inflammatory and T cell effector gene signature. Acquired T cell immunity and long-term protection were evidenced as increased immunodominant T cell clones after AVS100 treatment. Last, AVS100 showed no mutagenicity, toxicity, or adverse effects in preclinical good laboratory practice studies, part of the package that has led to US Food and Drug Administration clearance of an investigational new drug application for initiating clinical trials. This would be a first-in-human combination therapy of pembrolizumab with HDAC6 inhibition for locally advanced or metastatic solid tumors.
Figures



References
-
- Bondarev A. D., Attwood M. M., Jonsson J., Chubarev V. N., Tarasov V. V., Schioth H. B., Recent developments of HDAC inhibitors: Emerging indications and novel molecules. Br. J. Clin. Pharmacol. 87, 4577–4597 (2021). - PubMed
-
- Li M., Zhuang Y., Shan B., Analysis of expression and functions of histone deacetylase 6 (HDAC6). Methods Mol. Biol. 1436, 85–94 (2016). - PubMed
-
- Boyault C., Sadoul K., Pabion M., Khochbin S., HDAC6, at the crossroads between cytoskeleton and cell signaling by acetylation and ubiquitination. Oncogene 26, 5468–5476 (2007). - PubMed
-
- Li Y., Shin D., Kwon S. H., Histone deacetylase 6 plays a role as a distinct regulator of diverse cellular processes. FEBS J. 280, 775–793 (2013). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

