ATP citrate lyase is an essential player in the metabolic rewiring induced by PTEN loss during T-ALL development
- PMID: 39546747
- PMCID: PMC11999213
- DOI: 10.1182/bloodadvances.2024013762
ATP citrate lyase is an essential player in the metabolic rewiring induced by PTEN loss during T-ALL development
Abstract
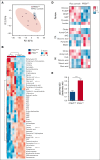
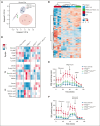
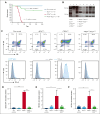
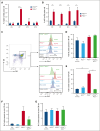
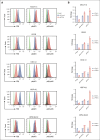
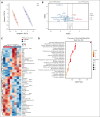
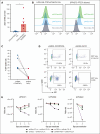
Alterations inactivating the tumor suppressor gene PTEN drive the development of solid and hematologic cancers, such as T-cell acute lymphoblastic leukemia (T-ALL), in which phosphatase and tensin homolog (PTEN) loss defines poor-prognosis patients. We investigated the metabolic rewiring induced by PTEN loss in T-ALL, aiming to identify novel metabolic vulnerabilities. We showed that the enzyme adenosine triphosphate (ATP) citrate lyase (ACLY) is strictly required for the transformation of thymic immature progenitors and the growth of human T-ALL, which remain dependent on ACLY activity even upon transformation. Although Pten-mutant mice all died within 17 weeks, the concomitant Acly deletion prevented disease initiation in 70% of the animals. In these animals, ACLY promoted B-cell lymphoma (BCL-2) epigenetic upregulation and prevented the apoptosis of premalignant double-positive thymocytes. Transcriptomic and metabolic analysis of primary T-ALL cells next translated our findings to the human pathology, showing that PTEN-altered T-ALL cells activate ACLY and are sensitive to its genetic targeting. ACLY activation thus represents a metabolic vulnerability with therapeutic potential for high-risk patients with T-ALL.
© 2025 American Society of Hematology. Published by Elsevier Inc. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures







References
-
- Suzuki A, Yamaguchi MT, Ohteki T, et al. T cell-specific loss of Pten leads to defects in central and peripheral tolerance. Immunity. 2001;14(5):523–534. - PubMed
-
- Tesio M, Trinquand A, Macintyre E, Asnafi V. Oncogenic PTEN functions and models in T-cell malignancies. Oncogene. 2016;35(30):3887–3896. - PubMed
-
- Tesio M, Trinquand A, Ballerini P, et al. Age-related clinical and biological features of PTEN abnormalities in T-cell acute lymphoblastic leukaemia. Leukemia. 2017;31(12):2594–2600. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

