STK19 facilitates the clearance of lesion-stalled RNAPII during transcription-coupled DNA repair
- PMID: 39547229
- PMCID: PMC12287594
- DOI: 10.1016/j.cell.2024.10.018
STK19 facilitates the clearance of lesion-stalled RNAPII during transcription-coupled DNA repair
Abstract
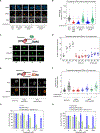
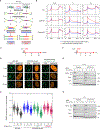
Transcription-coupled DNA repair (TCR) removes bulky DNA lesions impeding RNA polymerase II (RNAPII) transcription. Recent studies have outlined the stepwise assembly of TCR factors CSB, CSA, UVSSA, and transcription factor IIH (TFIIH) around lesion-stalled RNAPII. However, the mechanism and factors required for the transition to downstream repair steps, including RNAPII removal to provide repair proteins access to the DNA lesion, remain unclear. Here, we identify STK19 as a TCR factor facilitating this transition. Loss of STK19 does not impact initial TCR complex assembly or RNAPII ubiquitylation but delays lesion-stalled RNAPII clearance, thereby interfering with the downstream repair reaction. Cryoelectron microscopy (cryo-EM) and mutational analysis reveal that STK19 associates with the TCR complex, positioning itself between RNAPII, UVSSA, and CSA. The structural insights and molecular modeling suggest that STK19 positions the ATPase subunits of TFIIH onto DNA in front of RNAPII. Together, these findings provide new insights into the factors and mechanisms required for TCR.
Keywords: CSA; CSB; DNA repair; ELOF1; RNA polymerase II; STK19; TFIIH; UVSSA; nucleotide excision repair; transcription.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





Update of
-
STK19 facilitates the clearance of lesion-stalled RNAPII during transcription-coupled DNA repair.bioRxiv [Preprint]. 2024 Jul 22:2024.07.22.604575. doi: 10.1101/2024.07.22.604575. bioRxiv. 2024. Update in: Cell. 2024 Dec 12;187(25):7107-7125.e25. doi: 10.1016/j.cell.2024.10.018. PMID: 39091731 Free PMC article. Updated. Preprint.
References
-
- Mellon I, Spivak G, and Hanawalt PC (1987). Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell 51, 241–249. - PubMed
-
- Bohr VA, Smith CA, Okumoto DS, and Hanawalt PC (1985). DNA repair in an active gene: Removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell 40, 359–369. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

