The molecular chaperone ALYREF promotes R-loop resolution and maintains genome stability
- PMID: 39547511
- PMCID: PMC11665464
- DOI: 10.1016/j.jbc.2024.107996
The molecular chaperone ALYREF promotes R-loop resolution and maintains genome stability
Abstract
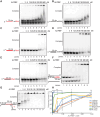
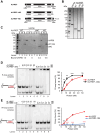
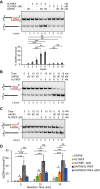
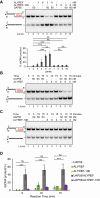
Unscheduled R-loops usually cause DNA damage and replication stress, and are therefore a major threat to genome stability. Several RNA processing factors, including the conserved THO complex and its associated RNA and DNA-RNA helicase UAP56, prevent R-loop accumulation in cells. Here, we investigate the function of ALYREF, an RNA export adapter associated with UAP56 and the THO complex, in R-loop regulation. We demonstrate that purified ALYREF promotes UAP56-mediated R-loop dissociation in vitro, and this stimulation is dependent on its interaction with UAP56 and R-loops. Importantly, we show that ALYREF binds DNA-RNA hybrids and R-loops. Consistently, ALYREF depletion causes R-loop accumulation and R-loop-mediated genome instability in cells. We propose that ALYREF, apart from its known role in RNA metabolism and export, is a key cellular R-loop coregulator, which binds R-loops and stimulates UAP56-driven resolution of unscheduled R-loops during transcription.
Keywords: ALYREF; DNA-RNA helicase; R-loop; TREX complex; UAP56; genome instability.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures

References
-
- Aguilera A., García-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol. Cell. 2012;46:115–124. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

