Vinculin-Arp2/3 interaction inhibits branched actin assembly to control migration and proliferation
- PMID: 39547716
- PMCID: PMC11568829
- DOI: 10.26508/lsa.202402583
Vinculin-Arp2/3 interaction inhibits branched actin assembly to control migration and proliferation
Abstract
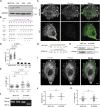
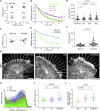
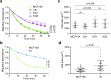
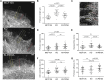
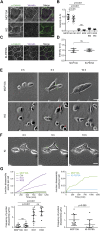
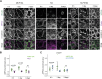
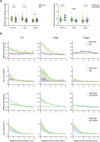
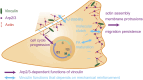
Vinculin is a mechanotransducer that reinforces links between cell adhesions and linear arrays of actin filaments upon myosin-mediated contractility. Both adhesions to the substratum and neighboring cells, however, are initiated within membrane protrusions that originate from Arp2/3-nucleated branched actin networks. Vinculin has been reported to interact with the Arp2/3 complex, but the role of this interaction remains poorly understood. Here, we compared the phenotypes of vinculin knock-out (KO) cells with those of knock-in (KI-P878A) cells, where the point mutation P878A that impairs the Arp2/3 interaction is introduced in the two vinculin alleles of MCF10A mammary epithelial cells. The interaction of vinculin with Arp2/3 inhibits actin polymerization at membrane protrusions and decreases migration persistence of single cells. In cell monolayers, vinculin recruits Arp2/3 and the vinculin-Arp2/3 interaction participates in cell-cell junction plasticity. Through this interaction, vinculin controls the decision to enter a new cell cycle as a function of cell density.
© 2024 James et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures










Similar articles
-
Talin-activated vinculin interacts with branched actin networks to initiate bundles.Elife. 2020 Nov 13;9:e53990. doi: 10.7554/eLife.53990. Elife. 2020. PMID: 33185186 Free PMC article.
-
Recruitment of the Arp2/3 complex to vinculin: coupling membrane protrusion to matrix adhesion.J Cell Biol. 2002 Dec 9;159(5):881-91. doi: 10.1083/jcb.200206043. Epub 2002 Dec 9. J Cell Biol. 2002. PMID: 12473693 Free PMC article.
-
Adhesive F-actin waves: a novel integrin-mediated adhesion complex coupled to ventral actin polymerization.PLoS One. 2011;6(11):e26631. doi: 10.1371/journal.pone.0026631. Epub 2011 Nov 1. PLoS One. 2011. PMID: 22069459 Free PMC article.
-
The Arp2/3 Regulatory System and Its Deregulation in Cancer.Physiol Rev. 2018 Jan 1;98(1):215-238. doi: 10.1152/physrev.00006.2017. Physiol Rev. 2018. PMID: 29212790 Review.
-
The stabilization of Arp2/3 complex generated actin filaments.Biochem Soc Trans. 2024 Feb 28;52(1):343-352. doi: 10.1042/BST20230638. Biochem Soc Trans. 2024. PMID: 38288872 Free PMC article. Review.
References
-
- Alexandrova AY, Arnold K, Schaub S, Vasiliev JM, Meister J-J, Bershadsky AD, Verkhovsky AB (2008) Comparative dynamics of retrograde actin flow and focal adhesions: Formation of nascent adhesions triggers transition from fast to slow flow. PLoS One 3: e3234. 10.1371/journal.pone.0003234 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
