Differentiation signals induce APOBEC3A expression via GRHL3 in squamous epithelia and squamous cell carcinoma
- PMID: 39548236
- PMCID: PMC11696371
- DOI: 10.1038/s44318-024-00298-9
Differentiation signals induce APOBEC3A expression via GRHL3 in squamous epithelia and squamous cell carcinoma
Abstract
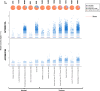
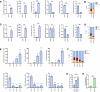
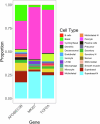
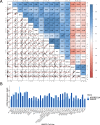
Two APOBEC DNA cytosine deaminase enzymes, APOBEC3A and APOBEC3B, generate somatic mutations in cancer, thereby driving tumour development and drug resistance. Here, we used single-cell RNA sequencing to study APOBEC3A and APOBEC3B expression in healthy and malignant mucosal epithelia, validating key observations with immunohistochemistry, spatial transcriptomics and functional experiments. Whereas APOBEC3B is expressed in keratinocytes entering mitosis, we show that APOBEC3A expression is confined largely to terminally differentiating cells and requires grainyhead-like transcription factor 3 (GRHL3). Thus, in normal tissue, neither deaminase appears to be expressed at high levels during DNA replication, the cell-cycle stage associated with APOBEC-mediated mutagenesis. In contrast, in squamous cell carcinoma we find that, there is expansion of GRHL3expression and activity to a subset of cells undergoing DNA replication and concomitant extension of APOBEC3A expression to proliferating cells. These findings suggest that APOBEC3A may play a functional role during keratinocyte differentiation, and offer a mechanism for acquisition of APOBEC3A mutagenic activity in tumours.
Keywords: APOBEC3A; Cancer Mutagenesis; GRHL3; HNSCC; Keratinocyte.
© 2024. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. TRF is an advisory board member of and holds stock options in APOBEC Discovery Ltd.
Figures




Update of
-
Differentiation signals induce APOBEC3A expression via GRHL3 in squamous epithelia and squamous cell carcinoma.Res Sq [Preprint]. 2024 Mar 4:rs.3.rs-3997426. doi: 10.21203/rs.3.rs-3997426/v1. Res Sq. 2024. Update in: EMBO J. 2025 Jan;44(1):1-29. doi: 10.1038/s44318-024-00298-9. PMID: 38496447 Free PMC article. Updated. Preprint.
References
-
- Allen-Hoffmann BL, Schlosser SJ, Ivarie CAR, Meisner LF, O’Connor SL, Sattler CA (2000) Normal growth and differentiation in a spontaneously immortalized near-diploid human keratinocyte cell line, NIKS. J Invest Dermatol 114:444–455 - PubMed
MeSH terms
Substances
Grants and funding
- M229/Rosetrees Trust (Rosetrees)
- CASE PhD studentship/AstraZeneca (AstraZeneca PLC)
- ACF-2023-26-009/National Institute for Health and Care Research (NIHR)
- CF-2021-2\101/Rosetrees Trust (Rosetrees)
- PhD2020\100002/Rosetrees Trust (Rosetrees)
- BB/V010271/2/UKRI | Biotechnology and Biological Sciences Research Council (BBSRC)
- NCI P01-CA234228/HHS | NIH | National Cancer Institute (NCI)
- P30 CA062203/CA/NCI NIH HHS/United States
- DRCNPG-Jun22\100004/Cancer Research UK (CRUK)
- R37 CA252081/CA/NCI NIH HHS/United States
- P01 CA234228/CA/NCI NIH HHS/United States
- CPRIT RR220053/Cancer Prevention and Research Institute of Texas (CPRIT)

