Multi-tissue metabolomic profiling reveals the crucial metabolites and pathways associated with scallop growth
- PMID: 39548384
- PMCID: PMC11566158
- DOI: 10.1186/s12864-024-11016-4
Multi-tissue metabolomic profiling reveals the crucial metabolites and pathways associated with scallop growth
Abstract
Background: Bivalves represent a vital economic resource in aquaculture for their high productivity and extensive market demand. Growth is one of the most important and desired aquaculture traits for bivalves, regulated by multiple levels, notably intricate metabolic processes. However, the understanding of the metabolic profiles that influence bivalve growth is limited, particularly from a multi-tissue perspective.
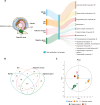
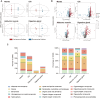
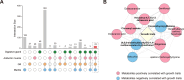
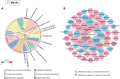
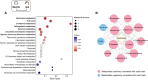
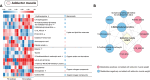
Results: In this study, metabolic profiles of multiple tissues of Chlamys farreri with different growth performance were systematically investigated by ultraperformance liquid chromatography quadrupole time-of-flight mass spectrometry (UPLC-Q-TOF-MS). Through comparing the metabolic variation between fast-growing (FG) scallops and slow-growing (SG) scallops, 613, 509, 105, and 192 significantly different metabolites (SDMs) were identified in the mantle, gill, adductor muscle, and digestive gland, respectively. Growth-related metabolic pathways including sphingolipid metabolism, fatty acid biosynthesis, and ABC transporter pathway, along with 11 SDMs associated with growth traits were identified in all four tissues, implying they were involved in the growth of multiple tissues in scallops. Tissue-specific metabolic profiling indicated that sulfur-containing amino acid metabolism in the mantle potentially contributed to shell growth, while the gill synergistically participated with the mantle through various metabolic processes, such as tyrosine metabolism, glycine, serine, and threonine metabolism and melanogenesis; energy metabolism was crucial for adductor muscle growth; and nutrients digestion and absorption in the digestive gland were linked to scallop growth.
Conclusions: Our results represent the first comprehensive analysis of the crucial pathways and metabolites associated with the growth of C. farreri, offering valuable insights for future bivalve aquaculture production.
Keywords: Chlamys farreri; Bivalve; Growth; Metabolomics; Multi-tissue.
© 2024. The Author(s).
Conflict of interest statement
Figures


Similar articles
-
Metabolomics comparison of metabolites and functional pathways in the gills of Chlamys farreri under cadmium exposure.Environ Toxicol Pharmacol. 2021 Aug;86:103683. doi: 10.1016/j.etap.2021.103683. Epub 2021 May 27. Environ Toxicol Pharmacol. 2021. PMID: 34052434
-
Dynamics of paralytic shellfish toxins and their metabolites during timecourse exposure of scallops Chlamys farreri and mussels Mytilus galloprovincialis to Alexandrium pacificum.Aquat Toxicol. 2018 Jul;200:233-240. doi: 10.1016/j.aquatox.2018.05.003. Epub 2018 May 16. Aquat Toxicol. 2018. PMID: 29778932
-
Molecular characterization of TGF-β type I receptor gene (Tgfbr1) in Chlamys farreri, and the association of allelic variants with growth traits.PLoS One. 2012;7(11):e51005. doi: 10.1371/journal.pone.0051005. Epub 2012 Nov 29. PLoS One. 2012. PMID: 23209843 Free PMC article.
-
Molecular characterization of RXR (Retinoid X Receptor) gene isoforms from the bivalve species Chlamys farreri.PLoS One. 2013 Sep 16;8(9):e74290. doi: 10.1371/journal.pone.0074290. eCollection 2013. PLoS One. 2013. PMID: 24066133 Free PMC article.
-
The interactions and biological pathways among metabolomics products of patients with coronary heart disease.Biomed Pharmacother. 2024 Apr;173:116305. doi: 10.1016/j.biopha.2024.116305. Epub 2024 Feb 29. Biomed Pharmacother. 2024. PMID: 38422653 Review.
Cited by
-
A Comprehensive and Comparative Study on the Biochemical Composition and Non-Volatile Taste Compounds of Thirteen Shellfish Species.Foods. 2025 Apr 30;14(9):1595. doi: 10.3390/foods14091595. Foods. 2025. PMID: 40361678 Free PMC article.
References
-
- FAO. Fishery and aquaculture statistics – yearbook 2020. FAO; 2023.
-
- Tan K, Ma H, Li S, Zheng H. Bivalves as future source of sustainable natural omega-3 polyunsaturated fatty acids. Food Chem. 2020;311:125907. - PubMed
-
- Venter L, Loots DT, Vosloo A, Van Jansen P, Lindeque JZ. Abalone growth and associated aspects: now from a metabolic perspective. Rev Aquacult. 2018;10:451–73.
-
- Guo L, Li L, Zhang S, Zhang G. Molecular cloning and characterization of the myostatin gene in a cultivated variety of bay scallop, Argopecten irradians. Aquaculture. 2012;350–353:192–9.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

