PNPLA3(148M) is a gain-of-function mutation that promotes hepatic steatosis by inhibiting ATGL-mediated triglyceride hydrolysis
- PMID: 39550037
- PMCID: PMC12164368
- DOI: 10.1016/j.jhep.2024.10.048
PNPLA3(148M) is a gain-of-function mutation that promotes hepatic steatosis by inhibiting ATGL-mediated triglyceride hydrolysis
Abstract
Background & aims: PNPLA3(148M) (patatin-like phospholipase domain-containing protein 3) is the most impactful genetic risk factor for steatotic liver disease. A key unresolved issue is whether PNPLA3(148M) confers a loss- or gain-of-function. Here we test the hypothesis that PNPLA3 causes steatosis by sequestering ABHD5 (α/β hydrolase domain-containing protein 5), the cofactor of ATGL (adipose TG lipase), thus limiting mobilization of hepatic triglyceride (TG).
Methods: We quantified and compared the physical interactions between ABHD5 and PNPLA3/ATGL in cultured hepatocytes using NanoBiT complementation assays and immunocytochemistry. Recombinant proteins purified from human cells were used to compare TG hydrolytic activities of PNPLA3 and ATGL in the presence or absence of ABHD5. Adenoviruses and adeno-associated viruses were used to express PNPLA3 in liver-specific Atgl-/- mice and to express ABHD5 in livers of Pnpla3M/M mice, respectively.
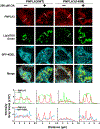
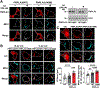
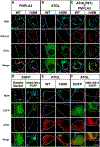
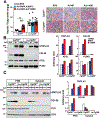
Results: ABHD5 interacted preferentially with PNPLA3 relative to ATGL in cultured hepatocytes. No differences were seen in the strength of the interactions between ABHD5 with PNPLA3(WT) and PNPLA3(148M). In contrast to prior findings, we found that PNPLA3, like ATGL, is activated by ABHD5 in in vitro assays using purified proteins. PNPLA3(148M)-associated inhibition of TG hydrolysis required that ATGL be expressed and that PNPLA3 be located on lipid droplets. Finally, overexpression of ABHD5 reversed the hepatic steatosis in Pnpla3M/M mice.
Conclusions: These findings support the premise that PNPLA3(148M) is a gain-of-function mutation that promotes hepatic steatosis by accumulating on lipid droplets and inhibiting ATGL-mediated lipolysis in an ABHD5-dependent manner. Our results predict that reducing, rather than increasing, PNPLA3 expression will be the best strategy to treat PNPLA3(148M)-associated steatotic liver disease.
Impact and implications: Steatotic liver disease (SLD) is a common complex disorder associated with both environmental and genetic risk factors. PNPLA3(148M) is the most impactful genetic risk factor for SLD and yet its pathogenic mechanism remains controversial. Herein, we provide evidence that PNPLA3(148M) promotes triglyceride (TG) accumulation by sequestering ABHD5, thus limiting its availability to activate ATGL. Although the substitution of methionine for isoleucine reduces the TG hydrolase activity of PNPLA3, the loss of enzymatic function is not directly related to the steatotic effect of the variant. It is the resulting accumulation of PNPLA3 on LDs that confers a gain-of-function by interfering with ATGL-mediated TG hydrolysis. These findings have implications for the design of potential PNPLA3(148M)-based therapies. Reducing, rather than increasing, PNPLA3 levels is predicted to reverse steatosis in susceptible individuals.
Keywords: Steatotic liver disease; lipid droplets; lipolysis; triglyceride hydrolase.
Copyright © 2024 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflict of interest Helen Hobbs is on the Corporate Board of Pfizer, Inc. Please refer to the accompanying ICMJE disclosure forms for further details.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

