Autoinflammatory patients with Golgi-trapped CDC42 exhibit intracellular trafficking defects leading to STING hyperactivation and ER stress
- PMID: 39550374
- PMCID: PMC11569173
- DOI: 10.1038/s41467-024-54294-y
Autoinflammatory patients with Golgi-trapped CDC42 exhibit intracellular trafficking defects leading to STING hyperactivation and ER stress
Abstract
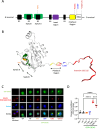
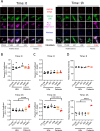
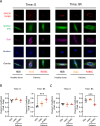
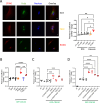
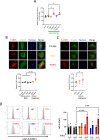
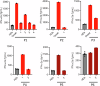
Most autoinflammatory diseases are caused by mutations in innate immunity genes. Previously, four variants in the RHO GTPase CDC42 were discovered in patients affected by syndromes generally characterized by neonatal-onset of cytopenia and auto-inflammation, including hemophagocytic lymphohistiocytosis and rash in the most severe form (NOCARH syndrome). However, the mechanisms responsible for these phenotypes remain largely elusive. Here, we show that the recurrent p.R186C CDC42 variant, which is trapped in the Golgi apparatus, elicits a block in both anterograde and retrograde transports. Consequently, it favours STING accumulation in the Golgi in a COPI-dependent manner. This is also observed for the other Golgi-trapped p.*192 C*24 CDC42 variant, but not for the p.Y64C and p.C188Y variants that do not accumulate in the Golgi. We demonstrate that the two Golgi-trapped CDC42 variants are the only ones that exhibit overactivation of the STING pathway and the type I interferon response, and elicit endoplasmic reticulum stress. Consistent with these results, patients carrying Golgi-trapped CDC42 mutants present very high levels of circulating IFNα at the onset of their disease. In conclusion, we report further mechanistic insights on the impact of the Golgi-trapped CDC42 variants. This increase in STING activation provides a rationale for combination treatments for these severe cases.
© 2024. The Author(s).
Conflict of interest statement
Figures


References
-
- Georgin-Lavialle, S. et al. Systemic autoinflammatory diseases: clinical state of the art. Best. Pract. Res. Clin. Rheumatol.34, 101529 (2020). - PubMed
-
- Aksentijevich, I. & Schnappauf, O. Molecular mechanisms of phenotypic variability in monogenic autoinflammatory diseases. Nat. Rev. Rheumatol.17, 405–425 (2021). - PubMed
-
- El Masri, R. & Delon, J. RHO GTPases: from new partners to complex immune syndromes. Nat. Rev. Immunol.21, 499–513 (2021). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

