Single cell and spatial transcriptomics highlight the interaction of club-like cells with immunosuppressive myeloid cells in prostate cancer
- PMID: 39550375
- PMCID: PMC11569175
- DOI: 10.1038/s41467-024-54364-1
Single cell and spatial transcriptomics highlight the interaction of club-like cells with immunosuppressive myeloid cells in prostate cancer
Abstract
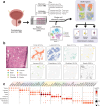
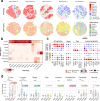
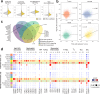
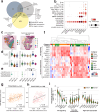
Prostate cancer treatment resistance is a significant challenge facing the field. Genomic and transcriptomic profiling have partially elucidated the mechanisms through which cancer cells escape treatment, but their relation toward the tumor microenvironment (TME) remains elusive. Here we present a comprehensive transcriptomic landscape of the prostate TME at multiple points in the standard treatment timeline employing single-cell RNA-sequencing and spatial transcriptomics data from 120 patients. We identify club-like cells as a key epithelial cell subtype that acts as an interface between the prostate and the immune system. Tissue areas enriched with club-like cells have depleted androgen signaling and upregulated expression of luminal progenitor cell markers. Club-like cells display a senescence-associated secretory phenotype and their presence is linked to increased polymorphonuclear myeloid-derived suppressor cell (PMN-MDSC) activity. Our results indicate that club-like cells are associated with myeloid inflammation previously linked to androgen deprivation therapy resistance, providing a rationale for their therapeutic targeting.
© 2024. The Author(s).
Conflict of interest statement
Figures

References
-
- Attard, G. et al. Prostate cancer. Lancet387, 70–82 (2016). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- SRA/PRJNA699369
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

