Exploring the impact of mitochondrial-targeting anthelmintic agents with GLUT1 inhibitor BAY-876 on breast cancer cell metabolism
- PMID: 39550554
- PMCID: PMC11568538
- DOI: 10.1186/s12885-024-13186-6
Exploring the impact of mitochondrial-targeting anthelmintic agents with GLUT1 inhibitor BAY-876 on breast cancer cell metabolism
Abstract
Background: Cancer cells alter their metabolic phenotypes with nutritional change. Single agent approaches targeting mitochondrial metabolism in cancer have failed due to either dose limiting off target toxicities, or lack of significant efficacy in vivo. To mitigate these clinical challenges, we investigated the potential utility of repurposing FDA approved mitochondrial targeting anthelmintic agents, niclosamide, IMD-0354 and pyrvinium pamoate, to be combined with GLUT1 inhibitor BAY-876 to enhance the inhibitory capacity of the major metabolic phenotypes exhibited by tumors.
Methods: To test this, we used breast cancer cell lines MDA-MB-231 and 4T1 which exhibit differing basal metabolic rates of glycolysis and mitochondrial respiration, respectively. Metabolic characterization was carried out using Seahorse XFe96 Bioanalyzer and statistical analysis was carried out via ANOVA.
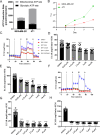
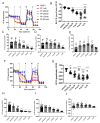
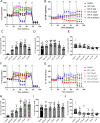
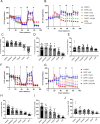
Results: Here, we found that specific responses to mitochondrial and glycolysis targeting agents elicit responses that correlate with tested cell lines basal metabolic rates and fuel preference, highlighting the potential to cater metabolism targeting treatment regimens based on specific tumor nutrient handling. Inhibition of GLUT1 with BAY-876 potently inhibited glycolysis in both MDA-MB-231 and 4T1 cells, and niclosamide and pyrvinium pamoate perturbed mitochondrial respiration that resulted in potent compensatory glycolysis in the cell lines tested.
Conclusion: In this regard, combination of BAY-876 with both mitochondrial targeting agents resulted in inhibition of compensatory glycolysis and subsequent metabolic crisis. These studies highlight targeting tumor metabolism as a combination treatment regimen that can be tailored by basal and compensatory metabolic phenotypes.
Keywords: Anthelmintic; Anticancer; BAY-876; Drug repurposing; Niclosamide; Pyrvinium.
© 2024. The Author(s).
Conflict of interest statement
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

