A Lassa virus live attenuated vaccine candidate that is safe and efficacious in guinea pigs
- PMID: 39551823
- PMCID: PMC11570604
- DOI: 10.1038/s41541-024-01012-w
A Lassa virus live attenuated vaccine candidate that is safe and efficacious in guinea pigs
Abstract
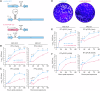
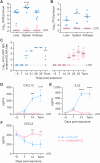
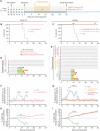
Lassa virus (LASV) is a rodent-borne mammarenavirus that causes tens to hundreds of thousands of human infections annually in Western Africa. Approximately 20% of these infections progress to Lassa fever (LF), an acute disease with case-fatality rates from ≈20-70%. Currently, there are no approved vaccines or specific therapeutics to prevent or treat LF. The LASV genome consists of a small (S) segment that has two genes, GP and NP, and a large (L) segment that has two genes, L and Z. In both segments, the two genes are separated by non-coding intergenic regions (IGRs). Recombinant LASVs (rLASVs), in which the L segment IGR was replaced with the S segment IGR or in which the GP gene was codon-deoptimized, lost fitness in vitro, were highly attenuated in vivo, and, when used as vaccines, protected domesticated guinea pigs from otherwise lethal LASV exposure. Here, we report the generation of rLASV/IGR-CD, which includes both determinants of attenuation and further enhances the safety of the vaccine compared with its predecessors. rLASV/IGR-CD grew to high titers in Vero cells, which are approved for human vaccine production, but did not cause signs of disease or pathology in guinea pigs. Importantly, guinea pigs vaccinated with rLASV/IGR-CD were completely protected from disease and death after a typically lethal exposure to wild-type LASV. Our data support the development of rLASV/IGR-CD as a live-attenuated LF vaccine with stringent safety features.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Figures



References
-
- Buckley, S. M., Casals, J. & Downs, W. G. Isolation and antigenic characterization of Lassa virus. Nature227, 174 (1970). - PubMed
-
- World Health Organization. R&D Blueprint. Lassa fever. https://www.who.int/teams/blueprint/lassa-fever (2018).
-
- Monath, T. P., Newhouse, V. F., Kemp, G. E., Setzer, H. W. & Cacciapuoti, A. Lassa virus isolation from Mastomys natalensis rodents during an epidemic in Sierra Leone. Science185, 263–265 (1974). - PubMed
Grants and funding
- 75N91019D00024/CA/NCI NIH HHS/United States
- R21AI121840/Division of Intramural Research, National Institute of Allergy and Infectious Diseases (Division of Intramural Research of the NIAID)
- R21 AI121840/AI/NIAID NIH HHS/United States
- U01 AI181212/AI/NIAID NIH HHS/United States
- R21AI169789/Division of Intramural Research, National Institute of Allergy and Infectious Diseases (Division of Intramural Research of the NIAID)
LinkOut - more resources
Full Text Sources
Miscellaneous

