DDX3 is critical for female fertility via translational control in oogenesis
- PMID: 39551844
- PMCID: PMC11570671
- DOI: 10.1038/s41420-024-02242-6
DDX3 is critical for female fertility via translational control in oogenesis
Abstract
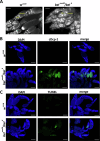
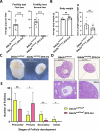
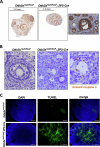
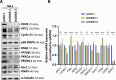
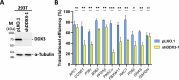
DEAD-box RNA helicase 3 (DDX3) and its homologs play a vital role in translation initiation by unwinding secondary structures of selected mRNAs. The human DDX3 gene is located on the sex chromosomes, so there are DDX3X and DDX3Y. DDX3X is ubiquitously expressed in almost all tissues and critical for embryonic development, whereas DDX3Y is only expressed in the testis and essential for male fertility. Drosophila belle (bel) is the single ortholog of DDX3, and mutations in bel cause male and female infertility. Using Drosophila bel mutants and Ddx3x conditional knockout (cKO) mice, we confirmed the pivotal role of DDX3 in female fertility and ovarian development. Drosophila bel mutants exhibited female infertility and immature egg chambers. Consistently, oocyte-specific Ddx3x knockout in mice resulted in female infertility and impaired oogenesis. We further found that immature egg chambers in Drosophila bel mutants and impaired follicular development in oocyte-specific Ddx3x cKO mice were caused by excessive apoptosis. We also identified a set of DDX3 target genes involved in oocyte meiosis and maturation and demonstrated that DDX3 is involved in their translation in human cells. Our results suggest that DDX3 is critical for female fertility via translational control in oogenesis.
© 2024. The Author(s).
Conflict of interest statement
Figures


References
-
- Chuang RY, Weaver PL, Liu Z, Chang TH. Requirement of the DEAD-Box protein ded1p for messenger RNA translation. Science. 1997;275:1468–71. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

