Dysrupted microbial tryptophan metabolism associates with SARS-CoV-2 acute inflammatory responses and long COVID
- PMID: 39551951
- PMCID: PMC11581176
- DOI: 10.1080/19490976.2024.2429754
Dysrupted microbial tryptophan metabolism associates with SARS-CoV-2 acute inflammatory responses and long COVID
Abstract
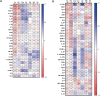
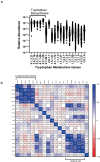
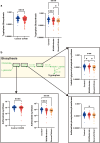
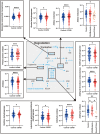
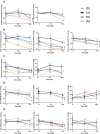
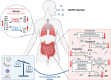
Protection against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection and risk of long COVID has been associated with the depletion or over-abundance of specific taxa within the gut microbiome. However, the microbial mechanisms mediating these effects are not yet known. We hypothesized that altered microbial production of tryptophan and its downstream derivatives might contribute to inappropriate immune responses to viral infection. In patients hospitalized with COVID-19 (n = 172), serum levels of tryptophan and indole-3-propionate (IPA) negatively correlated with serum levels of many proinflammatory mediators (including C-reactive protein and Serum amyloid A), while C-glycosyltryptophan (C-Trp), indole-3-lactic acid (ILA) and indole-3-acetic acid (IAA) levels were positively correlated with levels of acute phase proteins, proinflammatory cytokines, alarmins and chemokines. A similar pattern was observed in long COVID patients (n = 20) where tryptophan and IPA were negatively associated with a large number of serum cytokines, while C-Trp and IAA were positively associated with circulating cytokine levels. Metagenomic analysis of the fecal microbiota showed the relative abundance of genes encoding the microbial enzymes required for tryptophan production (e.g. anthranilate synthase) and microbial tryptophan metabolism was significantly lower in patients hospitalized with COVID-19 (n = 380) compared to healthy controls (n = 270). Microbial tryptophan metabolites reduced innate cell proinflammatory responses to cytosolic DNA sensor Stimulator of interferon genes (STING), toll-like receptor (TLR)-3 and TLR-4 stimulation in vitro, while IL-10 secretion was enhanced. Microbial tryptophan metabolites also modified ex vivo human lymphocyte responses by limiting the production of TH1 and TH17 associated cytokines, while enhancing secretion of IL-22. These data suggest that lower levels of tryptophan production and tryptophan metabolism by gut microbes may increase the risk of severe and chronic outcomes to SARS-CoV-2 infection due to impaired innate and adaptive responses to infection. Screening patients for lower-level microbiome capacity for tryptophan metabolism may help identify at-risk individuals.
Keywords: COVID-19; Microbiota; indoles; long COVID; tryptophan.
Conflict of interest statement
LOM is a consultant to PrecisionBiotics and has received research funding from GSK, Chiesi, Reckitt and Fonterra. LOM has participated in speaker’s bureau for Nestle, Nutricia, Yakult, Reckitt and Abbott. PWOT has received funding from Fonterra New Zealand and has participated in a speaker’s bureau for Yakult-Springer/Nature. None of the other authors report any potential conflict of interest.
Figures
References
-
- Levy M, Thaiss CA, Zeevi D, Dohnalová L, Zilberman-Schapira G, Mahdi JA, David E, Savidor A, Korem T, Herzig Y, et al. Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell. 2015. Dec 3;163(6):1428–1443. doi:10.1016/j.cell.2015.10.048. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
