Identification of copper metabolism-related markers in Parkinson's disease
- PMID: 39552415
- PMCID: PMC11574951
- DOI: 10.1080/07853890.2024.2425064
Identification of copper metabolism-related markers in Parkinson's disease
Abstract
Objectives: This study aimed to identify key genes related to copper metabolism in Parkinson's disease (PD), providing insight into their roles in disease progression.
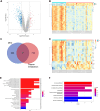
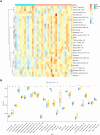
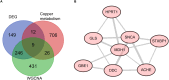
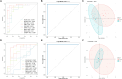
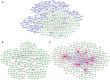
Methods: Using bioinformatic analyses, the study identified hub genes related to copper metabolism in PD patients. Differentially expressed genes (DEGs) were identified using the limma package, and copper-metabolism-related genes (CMRGs) were sourced from the Genecard database. Immune cell-related genes were derived through immune infiltration and Weighted Gene Co-expression Network Analysis (WGCNA). Hub genes were pinpointed by integrating DEGs, CMRGs, and immune cell-related genes. Functional analyses included Receiver Operating Characteristic (ROC) analysis, Ingenuity Pathway Analysis (IPA), and networks for miRNA-mRNA-transcription factor (TF), Competitive Endogenous RNA (ceRNA), and hub gene-drug interactions. Validation was performed in cerebrospinal fluid (CSF) samples from PD patients, while in vitro experiments utilized GBE1- overexpressing SH-SY5Y cells to examine cell proliferation, migration, and viability.
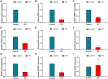
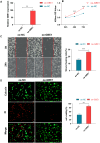
Results: Nine hub genes (HPRT1, GLS, SNCA, MDH1, GBE1, DDC, STXBP1, ACHE, and AGTR1) were identified from 753 CMRGs, 416 DEGs, and 951 immune cell-related genes. ROC analysis showed high predictive accuracy for PD, and principal component analysis (PCA) effectively distinguished PD patients from controls. IPA identified 20 significant pathways, and various networks highlighted miRNA, TF, and drug interactions with the hub genes. Hub gene expression was validated in PD CSF samples. GBE1-overexpressing cells displayed enhanced proliferation, migration, and viability.
Conclusions: The study identified nine copper metabolism-related genes as potential therapeutic targets in PD, highlighting their relevance in PD pathology and possible treatment pathways.
Keywords: GBE1; Parkinson’s disease; biomarker; copper metabolism; hub genes.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
