This is a preprint.
DDO-adjuvanted influenza A virus nucleoprotein mRNA vaccine induces robust humoral and cellular type 1 immune responses and protects mice from challenge
- PMID: 39553933
- PMCID: PMC11565765
- DOI: 10.1101/2024.10.27.620508
DDO-adjuvanted influenza A virus nucleoprotein mRNA vaccine induces robust humoral and cellular type 1 immune responses and protects mice from challenge
Update in
-
DDO-adjuvanted influenza A virus nucleoprotein mRNA vaccine induces robust humoral and cellular type 1 immune responses and protects mice from challenge.mBio. 2025 Feb 5;16(2):e0358924. doi: 10.1128/mbio.03589-24. Epub 2024 Dec 18. mBio. 2025. PMID: 39692514 Free PMC article.
Abstract
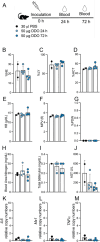
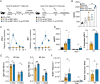
A challenge in viral vaccine development is to produce vaccines that generate both neutralizing antibodies to prevent infection and cytotoxic CD8+ T-cells that target conserved viral proteins and can eliminate infected cells to control virus spread. mRNA vaccines offer an opportunity to design vaccines based on conserved CD8-targeting epitopes, but achieving robust antigen-specific CD8+ T-cells remains a challenge. Here we tested the viral-derived oligonucleotide DDO268 as an adjuvant in the context of a model influenza A virus (IAV) nucleoprotein (NP) mRNA vaccine in C57BL/6 mice. DDO268 safely induced local type I interferon (IFN) production, stimulated dendritic cells type 1 (DC1) activation and migration to the draining lymph nodes, and improved the generation of IgG2c antibodies and antigen-specific effector Th1 CD4+ and CD8+ T-cells (IFNγ+TNFα+IL2+) when co-packaged with NP mRNA. The DDO268 adjuvanted vaccine provided enhanced protection against lethal IAV challenge and reduced the antigen dose required to achieve this protection. These results highlight the potential of DDO268 as an effective mRNA vaccine adjuvant and show that an IAV NP mRNA/DDO268 vaccine is a promising approach for generating protective immunity against conserved IAV epitopes.
Conflict of interest statement
Declaration of Competing Interest The following financial interests/personal relationships may be considered as potential competing interests: [The University of Pennsylvania and C.B.L. have a patent for Methods and Compositions for Stimulating Immune Response Using Potent Immunostimulatory RNA Motifs.].
Figures





References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
