This is a preprint.
A potently neutralizing and protective human antibody targeting antigenic site V on RSV and hMPV fusion glycoprotein
- PMID: 39554078
- PMCID: PMC11565947
- DOI: 10.1101/2024.10.31.621295
A potently neutralizing and protective human antibody targeting antigenic site V on RSV and hMPV fusion glycoprotein
Update in
-
A potently neutralizing and protective human antibody targeting antigenic site V on RSV and hMPV fusion glycoprotein.Cell Rep Med. 2026 Jan 16:102564. doi: 10.1016/j.xcrm.2025.102564. Online ahead of print. Cell Rep Med. 2026. PMID: 41547352
Abstract
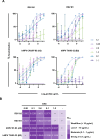
Human respiratory syncytial virus (RSV) and human metapneumovirus (hMPV) are frequent drivers of morbidity and mortality in susceptible populations, most often infantile, older adults, and immunocompromised. The primary target of neutralizing antibodies is the fusion (F) glycoprotein on the surface of the RSV and hMPV virion. As a result of the structural conservation between RSV and hMPV F, three antigenic regions are known to induce cross-neutralizing responses: sites III, IV, and V. Leveraging LIBRA-seq, we identify five RSV/hMPV cross-reactive human antibodies. One antibody, 5-1, potently neutralizes all tested viruses from the major subgroups of RSV and hMPV and provides protection against RSV and hMPV in a mouse challenge model. Structural analysis reveals that 5-1 utilizes an uncommon genetic signature to bind an epitope that spans sites Ø, II and V, defining a new mode of antibody cross-reactivity between RSV and hMPV F. These findings highlight the molecular and structural elements influencing RSV and hMPV cross-reactivity as well as the potential of antibody 5-1 for translational development.
Conflict of interest statement
DECELERATION OF INTERESTS A.A.A. and I.S.G. are listed as inventors on patents filed describing the antibodies discovered here. I.S.G. is listed as an inventor on patent applications for the LIBRA-seq technology. I.S.G. is a co-founder of AbSeek Bio. I.S.G. has served as a consultant for Sanofi. The Georgiev laboratory at VUMC has received unrelated funding from Merck and Takeda Pharmaceuticals. J.E.C. has served as a consultant for Luna Labs USA, Merck Sharp & Dohme Corporation, Emergent Biosolutions, a former member of the Scientific Advisory Boards of Gigagen (Grifols), of Meissa Vaccines, and BTG International, is founder of IDBiologics and receives royalties from UpToDate. The laboratory of J.E.C. received unrelated sponsored research agreements from AstraZeneca, Takeda Vaccines, and IDBiologics during the conduct of the study.
Figures



References
-
- Shi T., McAllister D.A., O’Brien K.L., Simoes E.A.F., Madhi S.A., Gessner B.D., Polack F.P., Balsells E., Acacio S., Aguayo C., et al. (2017). Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. The Lancet 390, 946–958. 10.1016/S0140-6736(17)30938-8. - DOI - PMC - PubMed
-
- O’Brien K.L., Baggett H.C., Brooks W.A., Feikin D.R., Hammitt L.L., Higdon M.M., Howie S.R.C., Deloria Knoll M., Kotloff K.L., Levine O.S., et al. (2019). Causes of severe pneumonia requiring hospital admission in children without HIV infection from Africa and Asia: the PERCH multi-country case-control study. The Lancet 394, 757–779. 10.1016/S0140-6736(19)30721-4. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
