This is a preprint.
Activating p53Y220C with a Mutant-Specific Small Molecule
- PMID: 39554093
- PMCID: PMC11565735
- DOI: 10.1101/2024.10.23.619961
Activating p53Y220C with a Mutant-Specific Small Molecule
Abstract
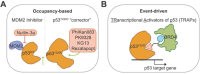
TP53 is the most commonly mutated gene in cancer, but it remains recalcitrant to clinically meaningful therapeutic reactivation. We present here the discovery and characterization of a small molecule chemical inducer of proximity that activates mutant p53. We named this compound TRanscriptional Activator of p53 (TRAP-1) due to its ability to engage mutant p53 and BRD4 in a ternary complex, which potently activates mutant p53 and triggers robust p53 target gene transcription. Treatment of p53Y220C expressing pancreatic cell lines with TRAP-1 results in rapid upregulation of p21 and other p53 target genes and inhibits the growth of p53Y220C-expressing cell lines. Negative control compounds that are unable to form a ternary complex do not have these effects, demonstrating the necessity of chemically induced proximity for the observed pharmacology. This approach to activating mutant p53 highlights how chemically induced proximity can be used to restore the functions of tumor suppressor proteins that have been inactivated by mutation in cancer.
Conflict of interest statement
Competing interests: All other authors declare no competing interests.
Figures


References
-
- Lavin M.F. & Gueven N. The complexity of p53 stabilization and activation. Cell Death Differ 13, 941–50 (2006). - PubMed
-
- Martincorena I. & Campbell P.J. Somatic mutation in cancer and normal cells. Science 349, 1483–9 (2015). - PubMed
-
- Olivier M. et al. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum Mutat 19, 607–14 (2002). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
