This is a preprint.
Phosphorylation of HP1/Swi6 relieves competition with Suv39/Clr4 on nucleosomes and enables H3K9 trimethyl spreading
- PMID: 39554105
- PMCID: PMC11565791
- DOI: 10.1101/2024.10.25.620326
Phosphorylation of HP1/Swi6 relieves competition with Suv39/Clr4 on nucleosomes and enables H3K9 trimethyl spreading
Abstract
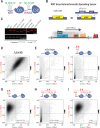
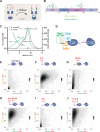
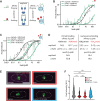
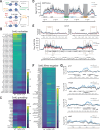
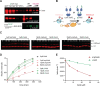
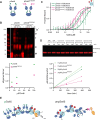
Heterochromatin formation in Schizosaccharomyces pombe requires the spreading of histone 3 (H3) Lysine 9 (K9) methylation (me) from nucleation centers by the H3K9 methylase, Suv39/Clr4, and the reader protein, HP1/Swi6. To accomplish this, Suv39/Clr4 and HP1/Swi6 have to associate with nucleosomes both nonspecifically, binding DNA and octamer surfaces and specifically, via recognition of methylated H3K9 by their respective chromodomains. However, how both proteins avoid competition for the same nucleosomes in this process is unclear. Here, we show that phosphorylation tunes oligomerization and the nucleosome affinity of HP1/Swi6 such that it preferentially partitions onto Suv39/Clr4's trimethyl product rather than its unmethylated substrates. Preferential partitioning enables efficient conversion from di-to trimethylation on nucleosomes in vitroand H3K9me3 spreading in vivo. Together, our data suggests that phosphorylation of HP1/Swi6 creates a regime that increases oligomerization and relieves competition with the "read-write" mechanism of Suv39/Clr4, together promoting for productive heterochromatin spreading.
Keywords: H3K9 trimethylation; HP1; heterochromatin spreading; phosphorylation; “reader” “writer” competition.
Conflict of interest statement
Competing interests The authors declare that they have no competing interests.
Figures

References
-
- Aagaard L, Schmid M, Warburton P, Jenuwein T. 2000. Mitotic phosphorylation of SUV39H1, a novel component of active centromeres, coincides with transient accumulation at mammalian centromeres. J Cell Sci 113 (Pt 5): 817–829. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
