This is a preprint.
Endogenous EWSR1-FLI1 degron alleles enable control of fusion oncoprotein expression in tumor cell lines and xenografts
- PMID: 39554175
- PMCID: PMC11566046
- DOI: 10.1101/2024.10.27.620498
Endogenous EWSR1-FLI1 degron alleles enable control of fusion oncoprotein expression in tumor cell lines and xenografts
Abstract
Pediatric malignancies frequently harbor chromosomal translocations that induce expression of fusion oncoproteins. The EWSR1-FLI1 fusion oncoprotein acts as a neomorphic transcription factor and is the dominant genetic driver of Ewing's sarcoma. Interrogation of the mechanisms by which EWSR1-FLI1 drives tumorigenesis has been limited by a lack of model systems to precisely and selectively control its expression in patient-derived cell lines and xenografts. Here, we report the generation of a panel of patient-derived EWS cell lines in which inducible protein degrons were engineered into the endogenous EWSR1-FLI1 locus. These alleles enabled rapid and efficient depletion of EWSR1-FLI1. Complete suppression of EWSR1-FLI1 induced a reversible cell cycle arrest at the G1-S checkpoint, and we identified a core set of transcripts downstream of EWSR1-FLI1 across multiple cell lines and degron systems. Additionally, depletion of EWSR1-FLI1 potently suppressed tumor growth in xenograft models validating efforts to directly target EWSR1-FLI1 in Ewing's sarcoma.
Keywords: EWSR1-FLI1; Ewing’s sarcoma; auxin-inducible degron; fusion oncoprotein; inducible degron; small molecule assisted shutoff; xenograft.
Conflict of interest statement
DECLARATION OF INTERESTS: The authors declare no competing interests
Figures


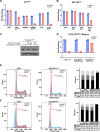
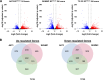

References
-
- Druker B.J., Sawyers C.L., Kantarjian H., Resta D.J., Reese S.F., Ford J.M., Capdeville R., and Talpaz M. (2001). Activity of a Specific Inhibitor of the BCR-ABL Tyrosine Kinase in the Blast Crisis of Chronic Myeloid Leukemia and Acute Lymphoblastic Leukemia with the Philadelphia Chromosome. N. Engl. J. Med. 344, 1038–1042. 10.1056/nejm200104053441402. - DOI - PubMed
-
- Druker B.J., Talpaz M., Resta D.J., Peng B., Buchdunger E., Ford J.M., Lydon N.B., Kantarjian H., Capdeville R., Ohno-Jones S., et al. (2001). Efficacy and Safety of a Specific Inhibitor of the BCR-ABL Tyrosine Kinase in Chronic Myeloid Leukemia. N. Engl. J. Med. 344, 1031–1037. 10.1056/nejm200104053441401. - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
