This is a preprint.
A Structural Mechanism for Noncanonical GPCR Signal Transduction in the Hedgehog Pathway
- PMID: 39554190
- PMCID: PMC11565934
- DOI: 10.1101/2024.10.31.621410
A Structural Mechanism for Noncanonical GPCR Signal Transduction in the Hedgehog Pathway
Abstract
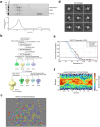
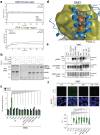
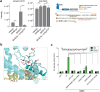
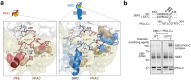
The Hedgehog (Hh) signaling pathway is fundamental to embryogenesis, tissue homeostasis, and cancer. Hh signals are transduced via an unusual mechanism: upon agonist-induced phosphorylation, the noncanonical G protein-coupled receptor SMOOTHENED (SMO) binds the catalytic subunit of protein kinase A (PKA-C) and physically blocks its enzymatic activity. By combining computational structural approaches with biochemical and functional studies, we show that SMO mimics strategies prevalent in canonical GPCR and PKA signaling complexes, despite little sequence or secondary structural homology. An intrinsically disordered region of SMO binds the PKA-C active site, resembling the PKA regulatory subunit (PKA-R) / PKA-C holoenzyme, while the SMO transmembrane domain binds a conserved PKA-C interaction hub, similar to other GPCR-effector complexes. In contrast with prevailing GPCR signal transduction models, phosphorylation of SMO promotes intramolecular electrostatic interactions that stabilize key structural elements within the SMO cytoplasmic domain, thereby remodeling it into a PKA-inhibiting conformation. Our work provides a structural mechanism for a central step in the Hh cascade and defines a paradigm for disordered GPCR domains to transmit signals intracellularly.
Figures













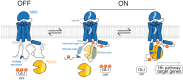
References
-
- Pierce K. L., Premont R. T. & Lefkowitz R. J. Seven-transmembrane receptors. Nat. Rev. Mol. Cell Biol. 3, 639–650 (2002). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
