The landscape of Arabidopsis tRNA aminoacylation
- PMID: 39555621
- PMCID: PMC11658184
- DOI: 10.1111/tpj.17146
The landscape of Arabidopsis tRNA aminoacylation
Abstract
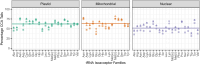
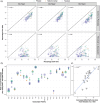
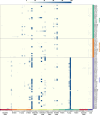
The function of transfer RNAs (tRNAs) depends on enzymes that cleave primary transcript ends, add a 3' CCA tail, introduce post-transcriptional base modifications, and charge (aminoacylate) mature tRNAs with the correct amino acid. Maintaining an available pool of the resulting aminoacylated tRNAs is essential for protein synthesis. High-throughput sequencing techniques have recently been developed to provide a comprehensive view of aminoacylation state in a tRNA-specific fashion. However, these methods have never been applied to plants. Here, we treated Arabidopsis thaliana RNA samples with periodate and then performed tRNA-seq to distinguish between aminoacylated and uncharged tRNAs. This approach successfully captured every tRNA isodecoder family and detected expression of additional tRNA-like transcripts. We found that estimated aminoacylation rates and CCA tail integrity were significantly higher on average for organellar (mitochondrial and plastid) tRNAs than for nuclear/cytosolic tRNAs. Reanalysis of previously published human cell line data showed a similar pattern. Base modifications result in nucleotide misincorporations and truncations during reverse transcription, which we quantified and used to test for relationships with aminoacylation levels. We also determined that the Arabidopsis tRNA-like sequences (t-elements) that are cleaved from the ends of some mitochondrial messenger RNAs have post-transcriptionally modified bases and CCA-tail addition. However, these t-elements are not aminoacylated, indicating that they are only recognized by a subset of tRNA-interacting enzymes and do not play a role in translation. Overall, this work provides a characterization of the baseline landscape of plant tRNA aminoacylation rates and demonstrates an approach for investigating environmental and genetic perturbations to plant translation machinery.
Keywords: Arabidopsis thaliana; aminoacylation; mitochondrial t‐elements; post‐transcriptional modifications; tRNAs.
© 2024 The Author(s). The Plant Journal published by Society for Experimental Biology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



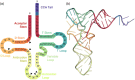

Similar articles
-
A robust method for measuring aminoacylation through tRNA-Seq.Elife. 2024 Jul 30;12:RP91554. doi: 10.7554/eLife.91554. Elife. 2024. PMID: 39076160 Free PMC article.
-
Combining tRNA sequencing methods to characterize plant tRNA expression and post-transcriptional modification.RNA Biol. 2021 Jan;18(1):64-78. doi: 10.1080/15476286.2020.1792089. Epub 2020 Jul 25. RNA Biol. 2021. PMID: 32715941 Free PMC article.
-
Determination of tRNA aminoacylation levels by high-throughput sequencing.Nucleic Acids Res. 2017 Aug 21;45(14):e133. doi: 10.1093/nar/gkx514. Nucleic Acids Res. 2017. PMID: 28586482 Free PMC article.
-
Controlling translation via modulation of tRNA levels.Wiley Interdiscip Rev RNA. 2015 Jul-Aug;6(4):453-70. doi: 10.1002/wrna.1287. Epub 2015 Apr 28. Wiley Interdiscip Rev RNA. 2015. PMID: 25919480 Free PMC article. Review.
-
The tRNA identity landscape for aminoacylation and beyond.Nucleic Acids Res. 2023 Feb 28;51(4):1528-1570. doi: 10.1093/nar/gkad007. Nucleic Acids Res. 2023. PMID: 36744444 Free PMC article. Review.
Cited by
-
Fungal-Derived tRNAs Are Expressed and Aminoacylated in Orchid Mitochondria.Mol Biol Evol. 2025 Feb 3;42(2):msaf025. doi: 10.1093/molbev/msaf025. Mol Biol Evol. 2025. PMID: 39882964 Free PMC article.
-
Nanopore sequencing of intact aminoacylated tRNAs.bioRxiv [Preprint]. 2024 Nov 18:2024.11.18.623114. doi: 10.1101/2024.11.18.623114. bioRxiv. 2024. PMID: 39605391 Free PMC article. Preprint.
References
-
- Arrivé, M. , Bruggeman, M. , Skaltsogiannis, V. , Coudray, L. , Quan, Y.‐F. , Schelcher, C. et al. (2023) A tRNA‐modifying enzyme facilitates RNase P activity in Arabidopsis nuclei. Nature Plants, 9(12), 2031–2041. - PubMed
-
- Betat, H. & Mörl, M. (2015) The CCA‐adding enzyme: a central scrutinizer in tRNA quality control. BioEssays, 37(9), 975–982. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

