The Emerging Roles of Multimolecular G-Quadruplexes in Transcriptional Regulation and Chromatin Organization
- PMID: 39555660
- PMCID: PMC11618987
- DOI: 10.1021/acs.accounts.4c00574
The Emerging Roles of Multimolecular G-Quadruplexes in Transcriptional Regulation and Chromatin Organization
Abstract
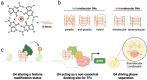
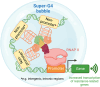
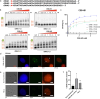
The ability of genomic DNA to adopt non-canonical secondary structures known as G-quadruplexes (G4s) under physiological conditions has been recognized for its potential regulatory function of various biological processes. Among those, transcription has recently emerged as a key process that can be heavily affected by G4 formation, particularly when these structures form at gene promoters. While the presence of G4s within gene promoters has been traditionally associated with transcriptional inhibition, in a model whereby G4s act as roadblocks to polymerase elongation, recent genomics experiments have revealed that the regulatory role of G4s in transcription is more complex than initially anticipated. Indeed, earlier studies linking G4-formation and transcription mainly relied on small-molecule ligands to stabilize and promote G4s, which might lead to disruption of protein-DNA interactions and local environments and, therefore, does not necessarily reflect the endogenous function of G4s at gene promoters. There is now strong evidence pointing toward G4s being associated with transcriptional enhancement, rather than repression, through multifaceted mechanisms such as recruitment of key transcriptional proteins, molding of chromatin architecture, and mode of phase separation. In this Account, we explore pivotal findings from our research on a particular subset of G4s, namely, those formed through interactions between distant genomic locations or independent nucleic acid strands, referred to as multimolecular G4s (mG4s), and discuss their active role in transcriptional regulation. We present our recent studies suggesting that the formation of mG4s may positively regulate transcription by inducing phase-separation and selectively recruiting chromatin-remodeling proteins. Our work highlighted how mG4-forming DNA and RNA sequences can lead to liquid-liquid phase separation (LLPS) in the absence of any protein. This discovery provided new insights into a potential mechanism by which mG4 can positively regulate active gene expression, namely, by establishing DNA networks based on distal guanine-guanine base pairing that creates liquid droplets at the interface of DNA loops. This is particularly relevant in light of the increasing evidence suggesting that G4 structures formed at enhancers can drive elevated expression of the associated genes. Given the complex three-dimensional nature of enhancers, our findings underscore how mG4 formation at enhancers would be particularly beneficial for promoting transcription. Moreover, we will elaborate on our recent discovery of a DNA repair and chromatin remodeling protein named Cockayne Syndrome B (CSB) that displays astonishing binding selectivity to mG4s over the more canonical unimolecular counterparts, suggesting another role of mG4s for molding chromatin architecture at DNA loops sites. Altogether, the studies presented in this Account suggest that mG4 formation in a chromatin context could be a crucial yet underexplored structural feature for transcriptional regulation. Whether mG4s actively regulate transcription or are formed as a mere consequence of chromatin plasticity remains to be elucidated. Still, given the novel insights offered by our research and the potential for mG4s to be selectively targeted by chemical and biological probes, we anticipate that further studies into the fundamental biology regulated by these structures can provide unprecedented opportunities for the development of therapeutic agents aimed at targeting nucleic acids from a fresh perspective.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
-
- Raguseo F.; Wang Y.; Li J.; Petrić Howe M.; Balendra R.; Huyghebaert A.; Vadukul D. M.; Tanase D. A.; Maher T. E.; Malouf L.; Rubio-Sánchez R.; Aprile F. A.; Elani Y.; Patani R.; Di Michele L.; Di Antonio M. The ALS/FTD-Related C9orf72 Hexanucleotide Repeat Expansion Forms RNA Condensates through Multimolecular G-Quadruplexes. Nat. Commun. 2023, 14 (1), 8272.10.1038/s41467-023-43872-1. - DOI - PMC - PubMed
-
- Robinson J.; Flint G.; Garner I.; Galli S.; Maher T. E.; Kuimova M. K.; Vilar R.; Mcneish I. A.; Brown R.; Keun H.; Antonio M. Di. G-Quadruplex Structures Regulate Long-Range Transcriptional Reprogramming to Promote Drug Resistance in Ovarian Cancer. bioRxiv 2024, 10.1101/2024.06.24.600010. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

