Securin regulates the spatiotemporal dynamics of separase
- PMID: 39556062
- PMCID: PMC11574863
- DOI: 10.1083/jcb.202312099
Securin regulates the spatiotemporal dynamics of separase
Abstract
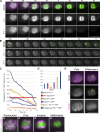
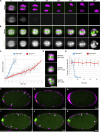
Separase regulates multiple aspects of the metaphase-to-anaphase transition. Separase cleaves cohesin to allow chromosome segregation and localizes to vesicles to promote exocytosis. The anaphase-promoting complex/cyclosome (APC/C) activates separase by ubiquitinating its inhibitory chaperone, securin, triggering its degradation. How this pathway controls the exocytic function of separase is unknown. During meiosis I, securin is degraded over several minutes, while separase rapidly relocalizes from kinetochore structures at the spindle and cortex to sites of action on chromosomes and vesicles at anaphase onset. The loss of cohesin coincides with the relocalization of separase to the chromosome midbivalent at anaphase onset. APC/C depletion prevents separase relocalization, while securin depletion causes precocious separase relocalization. Expression of non-degradable securin inhibits chromosome segregation, exocytosis, and separase localization to vesicles but not to the anaphase spindle. We conclude that APC/C-mediated securin degradation controls separase localization. This spatiotemporal regulation will impact the effective local concentration of separase for more precise targeting of substrates in anaphase.
© 2024 Sorensen Turpin et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures







Update of
-
Securin Regulates the Spatiotemporal Dynamics of Separase.bioRxiv [Preprint]. 2023 Dec 19:2023.12.12.571338. doi: 10.1101/2023.12.12.571338. bioRxiv. 2023. Update in: J Cell Biol. 2025 Feb 3;224(2):e202312099. doi: 10.1083/jcb.202312099. PMID: 38168402 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

