Polyamines mediate cellular energetics and lipid metabolism through mitochondrial respiration to facilitate virus replication
- PMID: 39556649
- PMCID: PMC11611256
- DOI: 10.1371/journal.ppat.1012711
Polyamines mediate cellular energetics and lipid metabolism through mitochondrial respiration to facilitate virus replication
Abstract
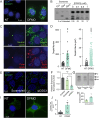
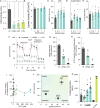
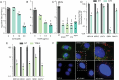
Polyamines are critical cellular components that regulate a variety of processes, including translation, cell cycling, and nucleic acid metabolism. The polyamines, putrescine, spermidine, and spermine, are found abundantly within cells and are positively-charged at physiological pH. Polyamine metabolism is connected to distinct other metabolic pathways, including nucleotide and amino acid metabolism. However, the breadth of the effect of polyamines on cellular metabolism remains to be fully understood. We recently demonstrated a role for polyamines in cholesterol metabolism, and following these studies, we measured the impact of polyamines on global lipid metabolism. We find that lipid droplets increase in number and size with polyamine depletion. We further demonstrate that lipid anabolism is markedly decreased, and lipid accumulation is due to reduced mitochondrial fatty acid oxidation. In fact, mitochondrial structure and function are largely ablated with polyamine depletion. To compensate, cells depleted of polyamines switch from aerobic respiration to glycolysis in a polyamine depletion-mediated Warburg-like effect. Finally, we show that inhibitors of lipid metabolism are broadly antiviral, suggesting that polyamines and lipids are promising antiviral targets. Together, these data demonstrate a novel role for polyamines in mitochondrial function, lipid metabolism, and cellular energetics.
Copyright: © 2024 Cruz-Pulido et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

