Disruption of the autism-associated Pcdh9 gene leads to transcriptional alterations, synapse overgrowth, and defective network activity in the CA1
- PMID: 39557582
- PMCID: PMC11638819
- DOI: 10.1523/JNEUROSCI.0491-24.2024
Disruption of the autism-associated Pcdh9 gene leads to transcriptional alterations, synapse overgrowth, and defective network activity in the CA1
Abstract
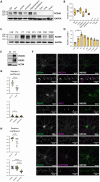
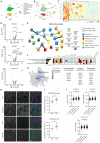
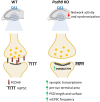
Protocadherins, a family of adhesion molecules with crucial role in cell-cell interactions, have emerged as key players in neurodevelopmental and psychiatric disorders. In particular, growing evidence links genetic alterations in Protocadherin 9 (PCDH9) gene with Autism Spectrum Disorder (ASD) and Major Depressive Disorder (MDD). Furthermore, Pcdh9 deletion induces neuronal defects in the mouse somatosensory cortex, accompanied by sensorimotor and memory impairment. However, the synaptic and molecular mechanisms of PCDH9 in the brain remain largely unknown, particularly concerning its impact on brain pathology. To address this question, we conducted a comprehensive investigation of PCDH9 role in the male mouse hippocampus at the ultrastructural, biochemical, transcriptomic, electrophysiological and network level. We show that PCDH9 mainly localizes at glutamatergic synapses and its expression peaks in the first week after birth, a crucial time window for synaptogenesis. Strikingly, Pcdh9 KO neurons exhibit oversized presynaptic terminal and postsynaptic density (PSD) in the CA1. Synapse overgrowth is sustained by the widespread up-regulation of synaptic genes, as revealed by single-nucleus RNA-seq (snRNA-seq), and the dysregulation of key drivers of synapse morphogenesis, including the SHANK2/CORTACTIN pathway. At the functional level, these structural and transcriptional abnormalities result into increased excitatory postsynaptic currents (mEPSC) and reduced network activity in the CA1 of Pcdh9 KO mice. In conclusion, our work uncovers Pcdh9 pivotal role in shaping the morphology and function of CA1 excitatory synapses, thereby modulating glutamatergic transmission within hippocampal circuits.Significance statement Converging evidence indicates that genetic alterations in Protocadherin 9 (PCDH9) gene are associated with Autism Spectrum Disorder (ASD) and Major Depressive Disorder (MDD). However, our understanding of PCDH9 physiological role and molecular mechanisms in the brain, as well as its connection to synaptic dysfunction and brain pathology, remains limited. Here we demonstrate that Pcdh9 regulates the transcriptional profile, morphology and function of glutamatergic synapses in the CA1, thereby tuning hippocampal network activity. Our results elucidate the molecular and synaptic mechanisms of a gene implicated in neurodevelopmental and psychiatric disorders, and suggest potential hippocampal alterations contributing to the cognitive deficits associated with these conditions.
Copyright © 2024 the authors.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
