Matching patients to clinical trials with large language models
- PMID: 39557832
- PMCID: PMC11574183
- DOI: 10.1038/s41467-024-53081-z
Matching patients to clinical trials with large language models
Abstract
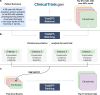
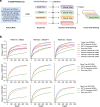
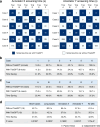
Patient recruitment is challenging for clinical trials. We introduce TrialGPT, an end-to-end framework for zero-shot patient-to-trial matching with large language models. TrialGPT comprises three modules: it first performs large-scale filtering to retrieve candidate trials (TrialGPT-Retrieval); then predicts criterion-level patient eligibility (TrialGPT-Matching); and finally generates trial-level scores (TrialGPT-Ranking). We evaluate TrialGPT on three cohorts of 183 synthetic patients with over 75,000 trial annotations. TrialGPT-Retrieval can recall over 90% of relevant trials using less than 6% of the initial collection. Manual evaluations on 1015 patient-criterion pairs show that TrialGPT-Matching achieves an accuracy of 87.3% with faithful explanations, close to the expert performance. The TrialGPT-Ranking scores are highly correlated with human judgments and outperform the best-competing models by 43.8% in ranking and excluding trials. Furthermore, our user study reveals that TrialGPT can reduce the screening time by 42.6% in patient recruitment. Overall, these results have demonstrated promising opportunities for patient-to-trial matching with TrialGPT.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Figures


Update of
-
Matching Patients to Clinical Trials with Large Language Models.ArXiv [Preprint]. 2024 Nov 18:arXiv:2307.15051v5. ArXiv. 2024. Update in: Nat Commun. 2024 Nov 18;15(1):9074. doi: 10.1038/s41467-024-53081-z. PMID: 37576126 Free PMC article. Updated. Preprint.
References
-
- Haddad, T. C. et al. Impact of a cognitive computing clinical trial matching system in an ambulatory oncology practice (American Society of Clinical Oncology, 2018).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

