Genomic and transcriptomic analyses identify distinctive features of triple-negative inflammatory breast cancer
- PMID: 39558017
- PMCID: PMC11574056
- DOI: 10.1038/s41698-024-00729-0
Genomic and transcriptomic analyses identify distinctive features of triple-negative inflammatory breast cancer
Abstract
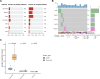
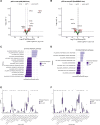
Triple-negative inflammatory breast cancer (TN-IBC) is the most aggressive type of breast cancer, yet its defining genomic, molecular, and immunological features remain largely unknown. In this study, we performed the largest and most comprehensive genomic and transcriptomic analyses of prospectively collected TN-IBC patient samples from a phase II clinical trial (ClinicalTrials.gov, NCT02876107, registered on August 22, 2016) and compared them to similarly analyzed stage III TN-non-IBC patient samples (ClinicalTrials.gov, NCT02276443, registered on October 21, 2014). We found that TN-IBC tumors have distinctive genomic, molecular, and immunological characteristics, including a lower tumor mutation load than TN-non-IBC, and an association of immunosuppressive tumor-infiltrating immune components with an unfavorable response to neoadjuvant chemotherapy. To our knowledge, this is the only study in which TN-IBC and TN-non-IBC samples were collected prospectively. Our analysis improves the understanding of the molecular landscape of the most aggressive subtype of breast cancer. Further studies are needed to discover novel prognostic biomarkers and druggable targets for TN-IBC.
© 2024. The Author(s).
Conflict of interest statement
Figures



References
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

