Salmonella cancer therapy metabolically disrupts tumours at the collateral cost of T cell immunity
- PMID: 39558103
- PMCID: PMC11628626
- DOI: 10.1038/s44321-024-00159-2
Salmonella cancer therapy metabolically disrupts tumours at the collateral cost of T cell immunity
Abstract
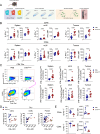
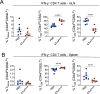
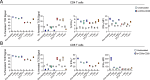
Bacterial cancer therapy (BCT) is a promising therapeutic for solid tumours. Salmonella enterica Typhimurium (STm) is well-studied amongst bacterial vectors due to advantages in genetic modification and metabolic adaptation. A longstanding paradox is the redundancy of T cells for treatment efficacy; instead, STm BCT depends on innate phagocytes for tumour control. Here, we used distal T cell receptor (TCR) and IFNγ reporter mice (Nr4a3-Tocky-Ifnγ-YFP) and a colorectal cancer (CRC) model to interrogate T cell activity during BCT with attenuated STm. We found that colonic tumour infiltrating lymphocytes (TILs) exhibited a variety of activation defects, including IFN-γ production decoupled from TCR signalling, decreased polyfunctionality and reduced central memory (TCM) formation. Modelling of T-cell-tumour interactions with a tumour organoid platform revealed an intact TCR signalosome, but paralysed metabolic reprogramming due to inhibition of the master metabolic controller, c-Myc. Restoration of c-Myc by deletion of the bacterial asparaginase ansB reinvigorated T cell activation, but at the cost of decreased metabolic control of the tumour by STm. This work shows for the first time that T cells are metabolically defective during BCT, but also that this same phenomenon is inexorably tied to intrinsic tumour suppression by the bacterial vector.
Keywords: Salmonella; Asparagine; Cancer Therapy; Immunometabolism; T Cells.
© 2024. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures










References
-
- André T, Shiu K-K, Kim TW, Jensen BV, Jensen LH, Punt C, Smith D, Garcia-Carbonero R, Benavides M, Gibbs P et al (2020) Pembrolizumab in microsatellite-instability–high advanced colorectal cancer. N. Engl J Med 383:2207–2218 - PubMed
-
- Bernal-Bayard J, Ramos-Morales F (2018) Molecular mechanisms used by Salmonella to evade the immune system. Curr Issues Mol Biol 25:133–168 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

