Functional and molecular insights into the role of Sae2 C-terminus in the activation of MRX endonuclease
- PMID: 39558159
- PMCID: PMC11662671
- DOI: 10.1093/nar/gkae1049
Functional and molecular insights into the role of Sae2 C-terminus in the activation of MRX endonuclease
Abstract
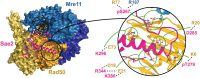
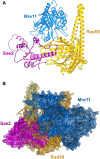
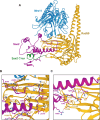
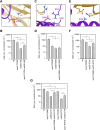
The yeast Sae2 protein, known as CtIP in mammals, once phosphorylated at Ser267, stimulates the endonuclease activity of the Mre11-Rad50-Xrs2 (MRX) complex to cleave DNA ends that possess hairpin structures or protein blocks, such as the Spo11 transesterase or trapped topoisomerases. Stimulation of the Mre11 endonuclease by Sae2 depends on a Rad50-Sae2 interaction, but the mechanism by which this is achieved remains to be elucidated. Through genetic studies, we show that the absence of the last 23 amino acids from the Sae2 C-terminus specifically impairs MRX-dependent DNA cleavage events, while preserving the other Sae2 functions. Employing AlphaFold3 protein structure predictions, we found that the Rad50-Sae2 interface involves not only phosphorylated Ser267 but also the phosphorylated Thr279 residue and the C-terminus of Sae2. This region engages in multiple interactions with residues that are mutated in rad50-s mutants, which are known to be specifically defective in the processing of Spo11-bound DNA ends. These interactions are critical for stabilizing the association between Sae2 and Rad50, thereby ensuring the correct positioning of Mre11 in its active endonucleolytic state.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

