OncoSplicing 3.0: an updated database for identifying RBPs regulating alternative splicing events in cancers
- PMID: 39558172
- PMCID: PMC11701682
- DOI: 10.1093/nar/gkae1098
OncoSplicing 3.0: an updated database for identifying RBPs regulating alternative splicing events in cancers
Abstract
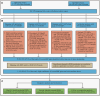
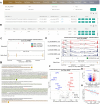
Alternative splicing (AS) is a crucial mechanism to regulate gene expression and protein complexity. RNA-binding proteins (RBPs) play an important role in regulating abnormal alternative splicing in cancers. However, few resources are available to identify specific RBPs responsible for regulating individual AS event. We have developed the OncoSplicing database for integrative analysis of clinically relevant alternative splicing events in TCGA cancers. Here, we further updated the OncoSplicing database by performing correlation analysis between the splicing and mRNA expression data from the TCGA cancers or GTEx tissues, mapping known RNA-binding motifs and eCLIP-seq peaks to all AS events, conducting splicing analysis for RNA-seq data from RBP perturbation experiments in the ENCODE project, and integrating exon and intron sequences for each AS event. With this updated database, users can easily identify potential RBPs responsible for the queried AS event and obtain sequences to design AS-specific primers and minigene constructs for experiment validation. Overall, compared to the previous version, the substantially updated OncoSplicing database (www.oncosplicing.com) offers a more valuable resource for users to identify RBPs responsible for regulating alternative splicing events in cancers.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
OncoSplicing: an updated database for clinically relevant alternative splicing in 33 human cancers.Nucleic Acids Res. 2022 Jan 7;50(D1):D1340-D1347. doi: 10.1093/nar/gkab851. Nucleic Acids Res. 2022. PMID: 34554251 Free PMC article.
-
CELF1 preferentially binds to exon-intron boundary and regulates alternative splicing in HeLa cells.Biochim Biophys Acta Gene Regul Mech. 2017 Sep;1860(9):911-921. doi: 10.1016/j.bbagrm.2017.07.004. Epub 2017 Jul 19. Biochim Biophys Acta Gene Regul Mech. 2017. PMID: 28733224
-
Emerging Multi-cancer Regulatory Role of ESRP1: Orchestration of Alternative Splicing to Control EMT.Curr Cancer Drug Targets. 2020;20(9):654-665. doi: 10.2174/1568009620666200621153831. Curr Cancer Drug Targets. 2020. PMID: 32564755
-
Coupling of alternative splicing and alternative polyadenylation.Acta Biochim Biophys Sin (Shanghai). 2024 Dec 3;57(1):22-32. doi: 10.3724/abbs.2024211. Acta Biochim Biophys Sin (Shanghai). 2024. PMID: 39632657 Free PMC article. Review.
-
RNA-Binding Protein-Mediated Alternative Splicing Regulates Abiotic Stress Responses in Plants.Int J Mol Sci. 2024 Sep 30;25(19):10548. doi: 10.3390/ijms251910548. Int J Mol Sci. 2024. PMID: 39408875 Free PMC article. Review.
Cited by
-
Multi-Omics Analysis of Survival-Related Splicing Factors and Identifies CRNKL1 as a Therapeutic Target in Esophageal Cancer.Genes (Basel). 2025 Mar 27;16(4):379. doi: 10.3390/genes16040379. Genes (Basel). 2025. PMID: 40282339 Free PMC article.
-
SurvDB: Systematic Identification of Potential Prognostic Biomarkers in 33 Cancer Types.Int J Mol Sci. 2025 Mar 20;26(6):2806. doi: 10.3390/ijms26062806. Int J Mol Sci. 2025. PMID: 40141449 Free PMC article.
References
-
- Matsuura K., Sawai H., Ikeo K., Ogawa S., Iio E., Isogawa M., Shimada N., Komori A., Toyoda H., Kumada T.et al. .. Genome-wide association study group for viral, genome-wide association study identifies TLL1 variant associated with development of hepatocellular carcinoma after eradication of hepatitis C virus infection. Gastroenterology. 2017; 152:1383–1394. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

