RAS signaling pathway is essential in regulating PIEZO1-mediated hepatic iron overload in dehydrated hereditary stomatocytosis
- PMID: 39558179
- PMCID: PMC11625994
- DOI: 10.1002/ajh.27523
RAS signaling pathway is essential in regulating PIEZO1-mediated hepatic iron overload in dehydrated hereditary stomatocytosis
Abstract
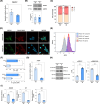
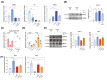
PIEZO1 encodes a mechanoreceptor, a cation channel activated by mechanical stimuli. Gain-of-function (GoF) variants in PIEZO1 cause dehydrated hereditary stomatocytosis (DHS), or xerocytosis, a pleiotropic syndrome characterized by anemia and iron overload. DHS patients develop hepatic iron overload independent of the degree of anemia and transfusion regimen. PIEZO1-GoF variants suppress hepcidin expression in both hepatic cellular model and constitutive/macrophage-specific Piezo1-GoF mice model. Therefore, PIEZO1-GoF variants regulate hepcidin expression by a crosstalk between hepatocytes (HCs) and macrophages with a still unknown mechanism. Transcriptomic and proteomics analysis in the human hepatic Hep3B cells engineered for the PIEZO1-R2456H variant (PIEZO1-KI) revealed alterations in the actin cytoskeleton regulation, MAPK cascade, and RAS signaling. These changes mainly occur through a novel key regulator, RRAS, whose protein and mRNA levels are regulated by PIEZO1 activation and inhibition. This regulation was further confirmed in C57BL/6 mouse primary HCs treated with Yoda-1 and/or GsMTx-4. Indeed, PIEZO1-KI cells exhibited hyper-activated RAS-GTPase activity that is rescued by PIEZO1 inhibition, restoring expression of the hepcidin gene HAMP. A negative correlation between RAS signaling and HAMP regulation was confirmed by inhibiting RAS-GTPase and MEK1-2 activity. Conversely, rescued HAMP gene expression requires downregulation of RRAS, confirming negative feedback between RAS-MAPK and BMP/SMADs pathways in HAMP regulation. We demonstrated that PIEZO1-GoF variants influence the actin cytoskeleton organization by activating the hepatic RAS signaling system. Understanding the role of RAS signaling in regulating iron metabolism could pave the way for new therapeutic strategies in DHS and other conditions characterized by iron overload.
© 2024 The Author(s). American Journal of Hematology published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no competing interests.
Figures


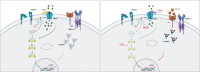
References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

