The STRING database in 2025: protein networks with directionality of regulation
- PMID: 39558183
- PMCID: PMC11701646
- DOI: 10.1093/nar/gkae1113
The STRING database in 2025: protein networks with directionality of regulation
Abstract
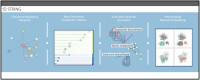
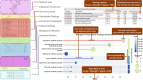
Proteins cooperate, regulate and bind each other to achieve their functions. Understanding the complex network of their interactions is essential for a systems-level description of cellular processes. The STRING database compiles, scores and integrates protein-protein association information drawn from experimental assays, computational predictions and prior knowledge. Its goal is to create comprehensive and objective global networks that encompass both physical and functional interactions. Additionally, STRING provides supplementary tools such as network clustering and pathway enrichment analysis. The latest version, STRING 12.5, introduces a new 'regulatory network', for which it gathers evidence on the type and directionality of interactions using curated pathway databases and a fine-tuned language model parsing the literature. This update enables users to visualize and access three distinct network types-functional, physical and regulatory-separately, each applicable to distinct research needs. In addition, the pathway enrichment detection functionality has been updated, with better false discovery rate corrections, redundancy filtering and improved visual displays. The resource now also offers improved annotations of clustered networks and provides users with downloadable network embeddings, which facilitate the use of STRING networks in machine learning and allow cross-species transfer of protein information. The STRING database is available online at https://string-db.org/.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Marsh J.A., Teichmann S.A.. Structure, dynamics, assembly, and evolution of protein complexes. Annu. Rev. Biochem. 2015; 84:551–575. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

