Integrated single-cell analysis reveals distinct epigenetic-regulated cancer cell states and a heterogeneity-guided core signature in tamoxifen-resistant breast cancer
- PMID: 39558215
- PMCID: PMC11572372
- DOI: 10.1186/s13073-024-01407-3
Integrated single-cell analysis reveals distinct epigenetic-regulated cancer cell states and a heterogeneity-guided core signature in tamoxifen-resistant breast cancer
Abstract
Background: Inter- and intra-tumor heterogeneity is considered a significant factor contributing to the development of endocrine resistance in breast cancer. Recent advances in single-cell RNA sequencing (scRNA-seq) and single-cell ATAC sequencing (scATAC-seq) allow us to explore inter- and intra-tumor heterogeneity at single-cell resolution. However, such integrated single-cell analysis has not yet been demonstrated to characterize the transcriptome and chromatin accessibility in breast cancer endocrine resistance.
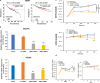
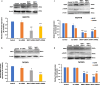
Methods: In this study, we conducted an integrated analysis combining scRNA-seq and scATAC-seq on more than 80,000 breast tissue cells from two normal tissues (NTs), three primary tumors (PTs), and three tamoxifen-treated recurrent tumors (RTs). A variety of cell types among breast tumor tissues were identified, PT- and RT-specific cancer cell states (CSs) were defined, and a heterogeneity-guided core signature (HCS) was derived through such integrated analysis. Functional experiments were performed to validate the oncogenic role of BMP7, a key gene within the core signature.
Results: We observed a striking level of cell-to-cell heterogeneity among six tumor tissues and delineated the primary to recurrent tumor progression, underscoring the significance of these single-cell level tumor cell clusters classified from scRNA-seq data. We defined nine CSs, including five PT-specific, three RT-specific, and one PT-RT-shared CSs, and identified distinct open chromatin regions of CSs, as well as a HCS of 137 genes. In addition, we predicted specific transcription factors (TFs) associated with the core signature and novel biological/metabolism pathways that mediate the communications between CSs and the tumor microenvironment (TME). We finally demonstrated that BMP7 plays an oncogenic role in tamoxifen-resistant breast cancer cells through modulating MAPK signaling pathways.
Conclusions: Our integrated single-cell analysis provides a comprehensive understanding of the tumor heterogeneity in tamoxifen resistance. We envision this integrated single-cell epigenomic and transcriptomic measure will become a powerful approach to unravel how epigenetic factors and the tumor microenvironment govern the development of tumor heterogeneity and to uncover potential therapeutic targets that circumvent heterogeneity-related failures.
Keywords: Breast tumor heterogeneity; Epigenetic-regulated cancer cell states; Single-cell analysis.
© 2024. The Author(s).
Conflict of interest statement
Figures





References
-
- Early Breast Cancer Trialists’ Collaborative G, Davies C, Godwin J, Gray R, Clarke M, Cutter D, Darby S, McGale P, Pan HC, Taylor C, et al. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet. 2011;378:771–84. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

