Ginkgetin Alleviates Inflammation and Senescence by Targeting STING
- PMID: 39558862
- PMCID: PMC11727237
- DOI: 10.1002/advs.202407222
Ginkgetin Alleviates Inflammation and Senescence by Targeting STING
Abstract
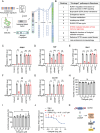
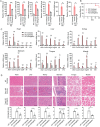
Ginkgo biloba extract is reported to have therapeutic effects on aging-related disorders. However, the specific component responsible for this biological function and its mechanism of action remain largely unknown. This study finds that Ginkgetin, an active ingredient of Ginkgo biloba extract, can alleviate cellular senescence and improve pathologies in multiple tissues of aging mice. To reveal the molecular mechanism of Ginkgetin's anti-aging effect, a graph convolutional network-based drug "on-target" pathway prediction algorithm for prediction is employed. The results indicate that the cGAS-STING pathway may be a potential target for Ginkgetin. Subsequent cell biological and biophysical data confirmed that Ginkgetin directly binds to the carboxy-terminal domain of STING protein, thereby inhibiting STING activation and signal transduction. Furthermore, in vivo pharmacodynamic data showed that Ginkgetin effectively alleviates systemic inflammation in Trex1-/- mice and inhibits the abnormally activated STING signaling in aging mouse model. In summary, this study, utilizing an artificial intelligence algorithm combined with pharmacological methods, confirms STING serves as a critical target for Ginkgetin in alleviating inflammation and senescence. Importantly, this study elucidates the specific component and molecular mechanism underlying the anti-aging effect of Ginkgo biloba extract, providing a robust theoretical basis for its therapeutic use.
Keywords: Ginkgetin; STING inhibitor; cGAS‐STING signaling; senescence; target identification.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
MeSH terms
Substances
Grants and funding
- T2225002/National Natural Science Foundation of China
- 82273855/National Natural Science Foundation of China
- 81903639/National Natural Science Foundation of China
- 82304379/National Natural Science Foundation of China
- 2023YFC2305904/National Key Research and Development Program of China
- 2022YFC3400504/National Key Research and Development Program of China
- 2023296/Youth Innovation Promotion Association CAS
- KF-202301/The open fund of state key laboratory of Pharmaceutical Biotechnology, Nanjing University, China
- 22ZR1474300/Natural Science Foundation of Shanghai
- E2G805H/SIMM-SHUTCM Traditional Chinese Medicine Innovation Joint Research Program
- 2023QNRC001/Young Elite Scientists Sponsorship Program by CAST
LinkOut - more resources
Full Text Sources
Medical
Research Materials
