Lactate and lysine lactylation of histone regulate transcription in cancer
- PMID: 39559217
- PMCID: PMC11570253
- DOI: 10.1016/j.heliyon.2024.e38426
Lactate and lysine lactylation of histone regulate transcription in cancer
Abstract
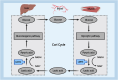
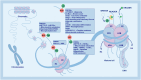
Histone lysine modifications were well-established epigenetic markers, with many types identified and extensively studied. The discovery of histone lysine lactylation had revealed a new form of epigenetic modification. The intensification of this modification was associated with glycolysis and elevated intracellular lactate levels, both of which were closely linked to cellular metabolism. Histone lactylation plays a crucial role in multiple cellular homeostasis, including immune regulation and cancer progression, thereby significantly influencing cell fate. Lactylation can modify both histone and non-histone proteins. This paper provided a comprehensive review of the typical epigenetic effects and lactylation on classical transcription-related lysine sites and summarized the known enzymes involved in histone lactylation and delactylation. Additionally, some discoveries of histone lactylation in tumor biology were also discussed, and some prospects for this field were put forward.
Keywords: Cancer; Histone; Lactate; Lysine lactylation; Post-translational modification; Transcription.
© 2024 Published by Elsevier Ltd.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
-
- Millán-Zambrano G., Burton A., Bannister A.J., Schneider R. Histone post-translational modifications - cause and consequence of genome function. Nat. Rev. Genet. 2022;23:563–580. - PubMed
-
- de la Cruz Muñoz-Centeno M., Millán-Zambrano G., Chávez S. A matter of packaging: influence of nucleosome positioning on heterologous gene expression. Methods Mol. Biol. 2012;824:51–64. - PubMed
-
- Levy B., Gibot S., Franck P., Cravoisy A., Bollaert P.-E. Relation between muscle Na+K+ ATPase activity and raised lactate concentrations in septic shock: a prospective study. Lancet. 2005;365:871–875. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

