Characterization of N6-methyladenosine long non-coding RNAs in sporadic congenital cataract and age-related cataract
- PMID: 39559306
- PMCID: PMC11528264
- DOI: 10.18240/ijo.2024.11.02
Characterization of N6-methyladenosine long non-coding RNAs in sporadic congenital cataract and age-related cataract
Abstract
Aim: To characterize the N6-methyladenosine (m6A) modification patterns in long non-coding RNAs (lncRNAs) in sporadic congenital cataract (CC) and age-related cataract (ARC).
Methods: Anterior capsule of the lens were collected from patients with CC and ARC. Methylated RNA immunoprecipitation with next-generation sequencing and RNA sequencing were performed to identify m6A-tagged lncRNAs and lncRNAs expression. Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses and Gene Ontology annotation were used to predict potential functions of the m6A-lncRNAs.
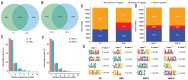
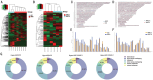
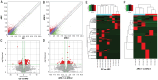
Results: Large amount of m6A peaks within lncRNA were identified for both CC and ARC, while the level was much higher in ARC (49 870 peaks) than that in CC (18 688 peaks), yet those difference between ARC in younger age group (ARC-1) and ARC in elder age group (ARC-2) was quite slight. A total of 1305 hypermethylated and 1178 hypomethylated lncRNAs, as well as 182 differential expressed lncRNAs were exhibited in ARC compared with CC. On the other hand, 5893 hypermethylated and 5213 hypomethylated lncRNAs, as well as 155 significantly altered lncRNA were identified in ARC-2 compared with ARC-1. Altered lncRNAs in ARC were mainly associated with the organization and biogenesis of intracellular organelles, as well as nucleotide excision repair.
Conclusion: Our results for the first time present an overview of the m6A methylomes of lncRNA in CC and ARC, providing a solid basis and uncovering a new insight to reveal the potential pathogenic mechanism of CC and ARC.
Keywords: N6-methyladenosine; RNA modification; age-related cataract; congenital cataract; epigenetics; long non-coding RNA.
International Journal of Ophthalmology Press.
Conflict of interest statement
Conflicts of Interest: Ye HF, None; Zhang X, None; Zhao ZN, None; Zheng C, None; Fei P, None; Xu Y, None; Lyu J, None; Chen JL, None; Guo XX, None; Zhu H, None; Zhao PQ, None.
Figures


Similar articles
-
Lens epithelium cell ferroptosis mediated by m6A-lncRNA and GPX4 expression in lens tissue of age-related cataract.BMC Ophthalmol. 2023 Dec 18;23(1):514. doi: 10.1186/s12886-023-03205-8. BMC Ophthalmol. 2023. PMID: 38110879 Free PMC article.
-
Identification and Characterization of N6-Methyladenosine CircRNAs and Methyltransferases in the Lens Epithelium Cells From Age-Related Cataract.Invest Ophthalmol Vis Sci. 2020 Aug 3;61(10):13. doi: 10.1167/iovs.61.10.13. Invest Ophthalmol Vis Sci. 2020. PMID: 32761139 Free PMC article.
-
m6A modification of lncRNA in middle ear cholesteatoma.Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2024 May 28;49(5):667-678. doi: 10.11817/j.issn.1672-7347.2024.230477. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2024. PMID: 39174880 Free PMC article.
-
Comprehensive analysis of m6A modification lncRNAs in high glucose and TNF-α induced human umbilical vein endothelial cells.Medicine (Baltimore). 2023 Mar 10;102(10):e33133. doi: 10.1097/MD.0000000000033133. Medicine (Baltimore). 2023. PMID: 36897718 Free PMC article.
-
Novel insights into mutual regulation between N6-methyladenosine modification and LncRNAs in tumors.Cancer Cell Int. 2023 Jun 26;23(1):127. doi: 10.1186/s12935-023-02955-1. Cancer Cell Int. 2023. PMID: 37365581 Free PMC article. Review.
Cited by
-
Human congenital cataract mutation in MYH9 alters F-actin organization and cell functions.Int J Ophthalmol. 2025 Jun 18;18(6):969-977. doi: 10.18240/ijo.2025.06.01. eCollection 2025. Int J Ophthalmol. 2025. PMID: 40534807 Free PMC article.
References
-
- Zhong YY, Zhou XE, Pan ZJ, Zhang JK, Pan J. Role of epigenetic regulatory mechanisms in age-related bone homeostasis imbalance. FASEB J. 2024;38(9):e23642. - PubMed
LinkOut - more resources
Full Text Sources
