A tight fit of the SIR dynamic epidemic model to daily cases of COVID-19 reported during the 2021-2022 Omicron surge in New York City: A novel approach
- PMID: 39559983
- PMCID: PMC11577707
- DOI: 10.1177/09622802241277956
A tight fit of the SIR dynamic epidemic model to daily cases of COVID-19 reported during the 2021-2022 Omicron surge in New York City: A novel approach
Abstract
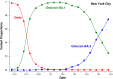
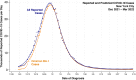
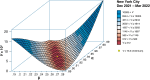
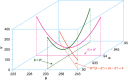
We describe a novel approach for recovering the underlying parameters of the SIR dynamic epidemic model from observed data on case incidence. We formulate a discrete-time approximation of the original continuous-time model and search for the parameter vector that minimizes the standard least squares criterion function. We show that the gradient vector and matrix of second-order derivatives of the criterion function with respect to the parameters adhere to their own systems of difference equations and thus can be exactly calculated iteratively. Applying our new approach, we estimated a four-parameter SIR model from daily reported cases of COVID-19 during the SARS-CoV-2 Omicron/BA.1 surge of December 2021-March 2022 in New York City. The estimated SIR model showed a tight fit to the observed data, but less so when we excluded residual cases attributable to the Delta variant during the initial upswing of the wave in December. Our analyses of both the real-world COVID-19 data and simulated case incidence data revealed an important problem of weak parameter identification. While our methods permitted for the separate estimation of the infection transmission parameter and the infection persistence parameter, only a linear combination of these two key parameters could be estimated with precision. The SIR model appears to be an adequate reduced-form description of the Omicron surge, but it is not necessarily the correct structural model. Prior information above and beyond case incidence data may be required to sharply identify the parameters and thus distinguish between alternative epidemic models.
Keywords: COVID-19; Newton–Raphson algorithm; Omicron variant; SARS-CoV-2; Susceptible-Infected-Removed model; heterogeneous mixing; inversion problem; nonlinear least squares; parameter identification; reduced form models; sloppy models; structural models; underreporting.
Conflict of interest statement
Declaration of conflicting interestsThe author declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures



References
-
- Kermack WO, McKendrick AG. A contribution to the mathematical theory of epidemics. Proce of the R Soc A: Math, Phys Eng Sci 1927; 115: 700–721.
-
- Magal P, Webb G. The parameter identification problem for SIR epidemic models: identifying unreported cases. J Math Biol 2018; 77: 1629–1648. - PubMed
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

