Staphylococcus epidermidis ST2 strains associated with bloodstream infections contain a unique mobile genetic element encoding a plasmin inhibitor
- PMID: 39560391
- PMCID: PMC11633098
- DOI: 10.1128/mbio.01907-24
Staphylococcus epidermidis ST2 strains associated with bloodstream infections contain a unique mobile genetic element encoding a plasmin inhibitor
Abstract
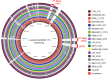
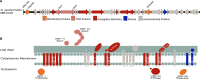
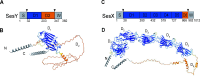
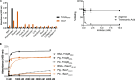
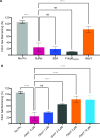
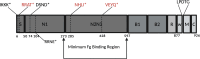
Staphylococcus epidermidis, a common commensal bacterium, is a leading cause of nosocomial catheter-associated bloodstream infections. S. epidermidis sequence type 2 (ST2) is specifically recognized globally for causing invasive disease. In this study, we identified a novel putative integrated conjugative element, pICE-Sepi-ST2, unique to the genomes of S. epidermidis ST2. Our investigation identified pICE-Sepi-ST2 in all ST2 isolates from bloodstream infections. Meanwhile, ST2 isolates from other infection sources, such as catheters, prosthetic joints, and fracture fixations, showed variable pICE-Sepi-ST2 prevalence. pICE-Sepi-ST2 encodes two putative cell wall anchored proteins that we have designated SesX and SesY. Biochemical characterization of SesY revealed that it binds both plasminogen (Plg) and plasmin (Pln) and inhibits Pln's ability to cleave a chromogenic substrate and degrade fibrin clots. Furthermore, all ST2 isolates containing a pICE-Sepi-ST2 also have a mutated sdrG gene. Thus, all ST2 isolates have two genetic modifications that target distinct steps in the hemostatic pathway. SdrG, which inhibits coagulation, is inactivated, and SesY, which inhibits fibrin, is introduced. These findings suggest that the hemostasis pathway is a strategic target for ST2 S. epidermidis bloodstream pathogenesis.
Importance: This study uncovers a new virulence mechanism in Staphylococcus epidermidis ST2 bloodstream isolates. We identify a mobile genetic element (MGE) characteristic of an integrated conjugated element (ICE). pICE-Sepi-ST2 carries the genetic information needed to produce a cell wall-anchored (CWA) protein called SesY. The results indicate that SesY binds to plasminogen (Plg) and plasmin (Pln) and inhibits Pln's degradation of fibrin clots. Genetic analysis showed that all ST2 bloodstream isolates can express the plasmin inhibitor SesY and carry a mutation in the SdrG gene, resulting in the expression of inactive SdrG. Thus, we describe a molecular pathway targeting the coagulation pathway that may be required for S. epidermidis ST2 to cause bloodstream infections.
Keywords: Staphylococcus epidermidis; bloodstream infections; coagulation pathway; integrated conjugated element; mobile genetic element; molecular pathogenesis; plasmin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Lee JYH, Monk IR, Gonçalves da Silva A, Seemann T, Chua KYL, Kearns A, Hill R, Woodford N, Bartels MD, Strommenger B, Laurent F, Dodémont M, Deplano A, Patel R, Larsen AR, Korman TM, Stinear TP, Howden BP. 2018. Global spread of three multidrug-resistant lineages of Staphylococcus epidermidis. Nat Microbiol 3:1175–1185. doi:10.1038/s41564-018-0230-7 - DOI - PMC - PubMed
-
- Shelburne SA, Dib RW, Endres BT, Reitzel R, Li X, Kalia A, Sahasrabhojane P, Chaftari A-M, Hachem R, Vargas-Cruz NS, Jiang Y, Garey K, Fowler VG Jr, Holland TL, Gu J, Miller W, Sakurai A, Arias CA, Aitken SL, Greenberg DE, Kim J, Flores AR, Raad I. 2020. Whole-genome sequencing of Staphylococcus epidermidis bloodstream isolates from a prospective clinical trial reveals that complicated bacteraemia is caused by a limited number of closely related sequence types. Clin Microbiol Infect 26:646. doi:10.1016/j.cmi.2019.10.008 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

